Abstract
Phlebotomine sandflies are important vectors of Leishmania spp. and phleboviruses causing disease in animals and humans. Morphological identification of phlebotomine sandflies to the species level is challenging, requiring microscopical examination of the genitalia, which is demanding and time consuming. Molecular sandfly species identification can be a practical solution to save resources since it enables further molecular studies capable of generating data, such as biting preferences by blood meal analysis. In this study, resorting to a sandfly dataset collected between 2014 and 2018 across Portuguese territory under active mosquito surveillance and sandfly specific surveys, we used molecular methods to explore the genetic diversity and spatial distribution, further exploring ecological co-variants of four sandfly species—Phlebotomus ariasi, P. perniciosus, P. sergenti, and Sergentomyia minuta—all of which are of public health importance. Sandflies were collected from Spring to Autumn (May–November) following local temperature patterns. P. perniciosus was the most widespread detected species, with a nationwide distribution. All studied species clustered together with known samples from the Iberian Peninsula. Further monitoring studies of sandfly species diversity, distribution, and seasonality are essential for surveillance and control of sandfly-borne pathogens both nationally and globally.
Keywords:
sandflies; Phlebotomus ariasi; P. perniciosus; P. sergenti; Sergentomyia minuta; COX1; Portugal 1. Introduction
Phlebotomine sandflies (Diptera, Psychodidae, and Phlebotominae) are small insects that can be found across the globe. They pose a threat from a public health point of view, since they serve as vectors of several human and animal pathogens, the most important of which are the parasites of the Leishmania genus (Kinetoplastida: Trypanosomatidae). Leishmaniasis is a disease caused by several species of Leishmania each transmitted by specific phlebotomine sandfly vectors [1]. In the Afro-Eurasia, sandflies of the genus Phlebotomus are proven vectors of human and canine leishmaniasis. In Portugal, visceral (VL) and cutaneous leishmaniasis (CL) caused by Leishmania infantum are endemic, with most reported cases being related to VL. This disease has so far been predominantly detected in children, but since the 1990s most cases are reported in HIV-infected adults [2]. Domesticated dogs (Canis lupus familiaris) are recognized as the main host and reservoir [3,4], but a recent increase in reported infections in cats (Felis catus) is questioning the role of this species in Leishmania’s zoonotic cycle in urban and peri-urban habitats [5,6,7,8]. The phlebotomine species associated with Leishmania transmission in Portugal are Phlebotomus perniciosus and P. ariasi [9].
In the Mediterranean region, in addition to Leishmania parasites, sandflies are currently responsible for the transmission of viruses of the Phlebovirus genus (Phenuiviridae), which includes 66 viral species [10]. Although the pathogenic potential of most Phenuiviridae is unknown, some are important disease agents, such as, e.g., the Rift Valley Fever phlebovirus (transmitted by mosquitoes) and the Toscana phlebovirus (TOSV) transmitted by sandflies. In Mediterranean countries, Naples and Sicilian phleboviruses are recognized as important disease agents causing mild febrile syndromes, commonly designated as “sandfly or three-day fever” [11]. TOSV is nonetheless considered the most relevant pathogen sandfly-borne phlebovirus, which can cause severe and potentially fatal neuro-invasive disease, responsible for a significant number of neurological disease cases during the summertime [12]. TOSV, Massillia phlebovirus (namely, Arrábida virus and Massillia virus isolates 21, 70, 127, 130) [13,14,15] and Alcube phlebovirus [14] are known to circulate in Portugal, but human infections have only been associated to TOSV and Sicilian phleboviruses [16].
There are almost 1000 species of sandflies distributed within six genera; however, only three genera, Phlebotomus, Lutzomyia, and Sergentomyia, have hematophagous species. The genera Phlebotomus and Sergentomyia are confined to Afro-Eurasia, and Lutzomyia species are found only in the Americas [17]. In Portugal, five sandfly species have so far been reported. P. papatasi (Scopoli, 1786) was firstly reported in 1913 [18], followed by the report in 1918 of P. perniciosus (Newstead, 1911) and P. sergenti (Parrot, 1917) [19]. P. ariasi (Tonnoir, 1921) and S. minuta (Rondani, 1843) were reported in 1944 by Meira and Ferreira [20]. Phlebotomine sandflies are fragile, tiny (1.5–3.5 mm), ectothermic, hairy insects that are unable to colonize new distant areas either by passive transport or individual motility, none being considered so far as invasive species [21]. Sandfly-borne pathogen transmission in endemic areas is strongly seasonal, being driven by environmental factors that affect vector and reservoir host abundance [21]. Adult female sandflies from hematophagous species feed on a wide range of mammals and birds. The sandflies’ life cycle takes at least six weeks to complete, with their vector activity being seasonal and linked to climate natural fluctuations, especially temperature and precipitation [22]. Depending on latitude, sandflies tend to be detected from April to November in Europe [22]. Phlebotomine sandflies’ species and inherent genetic lineages have diverse endurance to disparate climate conditions, with P. ariasi reported to be associated with cooler and humid environments, while P. perniciosus appears adapted to a wider range of climatic conditions [23].
Due to their small size, morphological identification of phlebotomine sandflies to species level requires meticulous microscopical examination of internal structures. DNA sequence data have been proven as a successful approach for sandfly species identification [24] with the advantage of enabling further molecular and population genetics studies. Investing on molecular approaches also has the advantage of providing research paths beyond species classification, such as characterizing biting preferences by blood meal analysis. Although several local reports of sandflies in Portugal are available, mainly related to reported Leishmania foci [9,20,24,25,26,27,28], country-wide studies of the population genetics of these insects are needed to better understand their distribution, population structure, and potential contribution for vector-borne pathogen transmission into human populations.
In this study, resorting to a sandfly dataset collected between 2014 and 2018 across Portuguese territory under active-mosquito- and sandfly-specific surveillance surveys, we explored the genetic diversity, spatial distribution, and ecological co-variates of four sandfly species—P. ariasi, P. perniciosus, P. sergenti, and S. minuta—all of which are of public health importance, both nationally and globally.
2. Materials and Methods
A total of 173 sandflies analysed in this study were collected using different methodologies: (1) specific for sandflies at their recognized habitats and (2) in mosquito surveillance directed collections. From 2014 to 2018, sandflies collected under the regular adult mosquitoes monitoring activities of the national REVIVE vector surveillance network [29] using CDC miniature light traps (John W Hock Company, Gainesville, FL, USA) baited mostly with dry ice or by aspiration were included in this study. Between May and October 2018 specific sandflies entomological field surveys took place in the Arrábida region (Palmela, Setúbal, and Sesimbra counties) and eastern Algarve (Loulé, Faro, Olhão, and Tavira counties), near or in animal facilities such as kennels, rabbit hutches, pigsties, chicken pens, or sheep corrals (using modified CDC miniature light traps with a reduced collection cup mesh size) and baited with dry ice. For vector samples with an identified presence of sandflies, collection dates, geographical coordinates (latitude and longitude), the altitude of collection sites and the capture methodology are shown in Table S1. When the altitude was not recorded in place, an estimation was made superimposing geographical coordinates with local topographic maps [30]. All samples were collected at public and private proprieties with the respective accountable owners’ knowledge and permission.
Alive sandflies were immediately processed or stored at −80 °C. Samples for DNA extraction were selected individually according to date, trapping origin, and gender. Samples were grinded with a mortar and pestle with liquid nitrogen and 500 µL of minimal essential medium supplied with 10% FBS, streptomycin (0.1 mg/mL), and amphotericin B (1 mg/mL). An aliquot of 300 µL was preserved at −80 °C, and the remaining volume was further grinded with 300 µL of Lysis Buffer (NUCLISENS® easyMAG, Biomérieux, Marcy-l’Étoile, France), added to the homogenizer cartridge (Invitrogen), and centrifuged at 12,000× g for 2 min to remove cellular debris and reduce lysate viscosity. Total nucleic acid extraction was performed using the prepared lysate suspensions in the automated platform NUCLISENS® easyMAG (Biomérieux).
Samples were molecularly identified using cytochrome c oxidase subunit I gene (COX1) of mitochondrial DNA using primers LCO 1490 and HCO 2198 [31], as previously described [32]. Vertebrate hosts constituting bloodmeals were identified by PCR amplification of a 508 bp fragment of CytB using primers MamC+ and MamC- [33] using DNA extracted from single blood-fed female sandflies. Bloodmeal analysis was performed by screening 39 engorged females belonging to the four identified species (11 for P. ariasi, 22 for P. perniciosus, 1 for P. sergenti, and 4 for S. minuta) (see Table S1). High Fidelity PCR Master (Roche, Mannheim, Germany) was used for PCR amplification according to the manufacturer’s instructions. Amplicons were visualized by agarose gel electrophoresis and purifed using Jet Quick-PCR purifcation kit (Genomed, Warszawa, Poland). Purified DNA fragments were sequenced in an ABI automated DNA capillary system (Applied Biosystems, Foster City, CA, USA) using the ABI Big Dye Terminator kit (Applied Biosystems). Sequences were assembled using the BioEdit software (version 7.2.5) [34]. Positive identification and host species assignment were made by sequence similarity searches (basic local alignment search tool, BLASTN algorithm, NCBI [35]. Positive (of known-origin animal blood) and negative controls were processed and correctly identified with the above-mentioned procedure. A bloodmeal was classified as mixed when chromatograms from each PCR demonstrated double-nucleotide peaks.
Partial COX1 sequences were submitted to the GenBank database (accession numbers MH559420–MH559497 and OL364747–L364845). Nucleotide consensus sequences of COX1 gene were aligned against multiple sequences available at GenBank for each species, using BioEdit. Maximum likelihood analysis was performed with a Tamura-3-parameter model with a gamma-distributed (G) rate among sites for P. ariasi and P. sergenti trees, a Tamura-3-parameter model for P. perniciosus, and a Tamura-Nei model with G rate among sites for S. minuta (the determined best fit models) using Molecular Evolutionary Genetics Analysis (MEGAX) software [36]. The robustness of the internal nodes was tested by 500 bootstrap replications. The resulting trees were then visualized and annotated with ggtree in R [37]. Metrics of genetic diversity, including the number of polymorphic sites and nucleotide diversity for COX1 sequences, were estimated using DnaSP v.5.0 [38].
The local temperature and the land type of each sample were obtained from Copernicus satellite data sources. For the land type, the data source used was “Land cover classification gridded maps from 1992 to present derived from satellite observations” [39] and for temperature it was “Essential climate variables for assessment of climate variability” [40]. We used each sample’s geo-location (closest latitude–longitude match between sample and satellite data) and the month and the year of collection to extract the temperature and the land type. Temperature was at a time resolution of mean per month per year, and land type was at a resolution of years. Land-type data came with 22 classifications (see [39]) and was here aggregated into five relevant classes: bare (class 200), cropland vegetation (classes 10, 11, 12, and 20), natural vegetation (classes 60, 70, 90, 100, 110, 120, 122, 130, and 180), mixed vegetation (i.e., natural and cropland; classes 30 and 40) and urban (class 190). The approach was the same as in our recent study in Portugal [41].
Portuguese districts (total of 18) were aggregated into five macro regions of interest: North (districts: Viana do Castelo, Braga, Porto, Vila Real, and Bragança); Central (Aveiro, Viseu, Guarda, Coimbra, Castelo Branco, Coimbra, and Leiria); Lisbon (Lisboa, Santarém, and Setúbal); Alentejo (Portalegre, Évora, and Beja); Algarve (Faro).
3. Results
A total of 173 sandflies, collected in 66 geographical sites in 46 counties and belonging to 16 districts in mainland Portugal (only missing data from two northern districts, Braga and Vila Real) from 2014 to 2018 were analysed in this study (Table S1). Molecular analysis of COX1 gene allowed the identification of 33 P. ariasi, 124 P. perniciosus, 3 P. sergenti, and 13 S. minuta. Below we explore and summarize the genetic population structure of these samples in the context of other already-known genetic sequences, as well as the seasonal timing of the different species, their geographical distributions, and ecological contexts.
3.1. Genetic Analysis
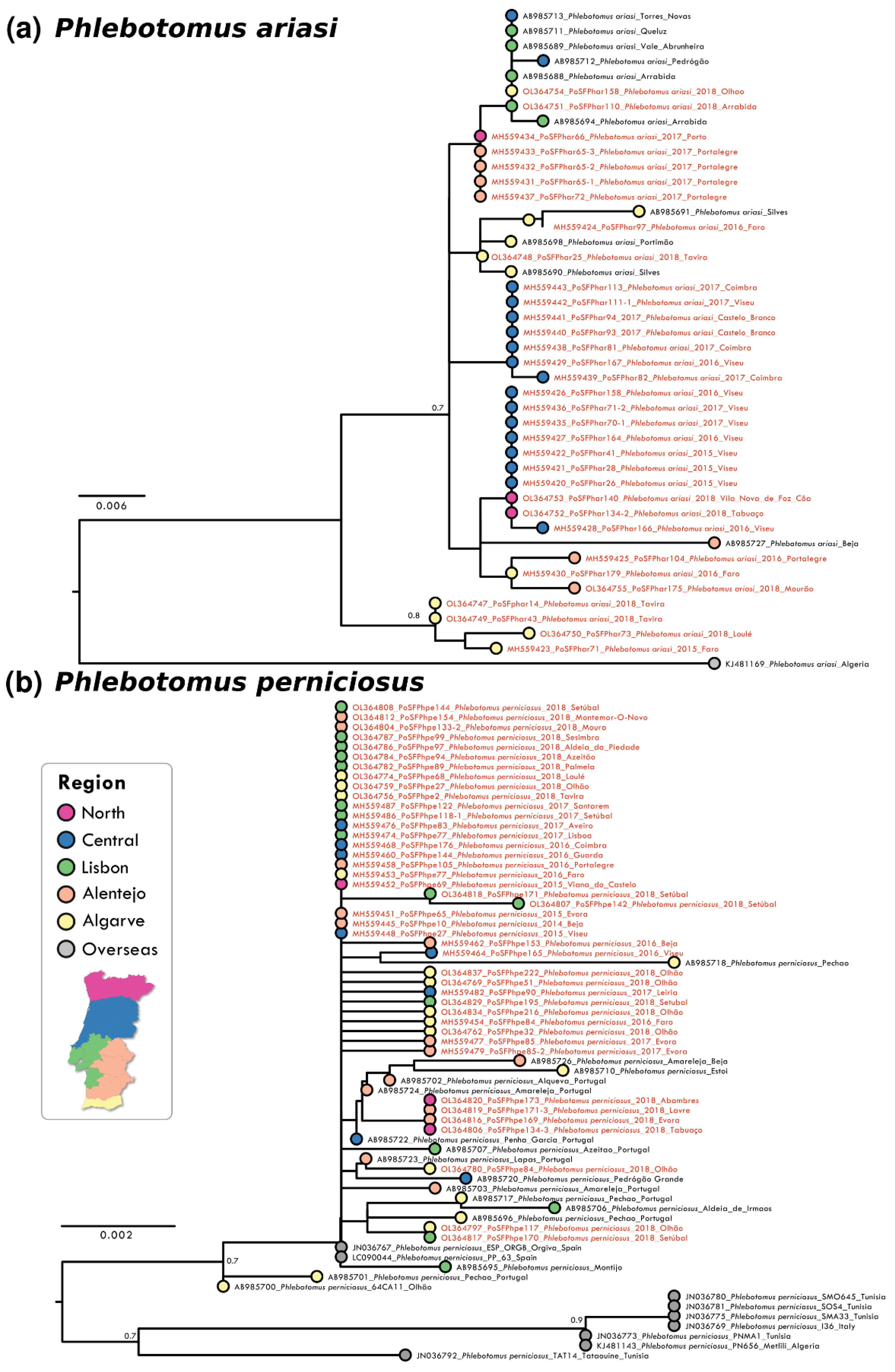
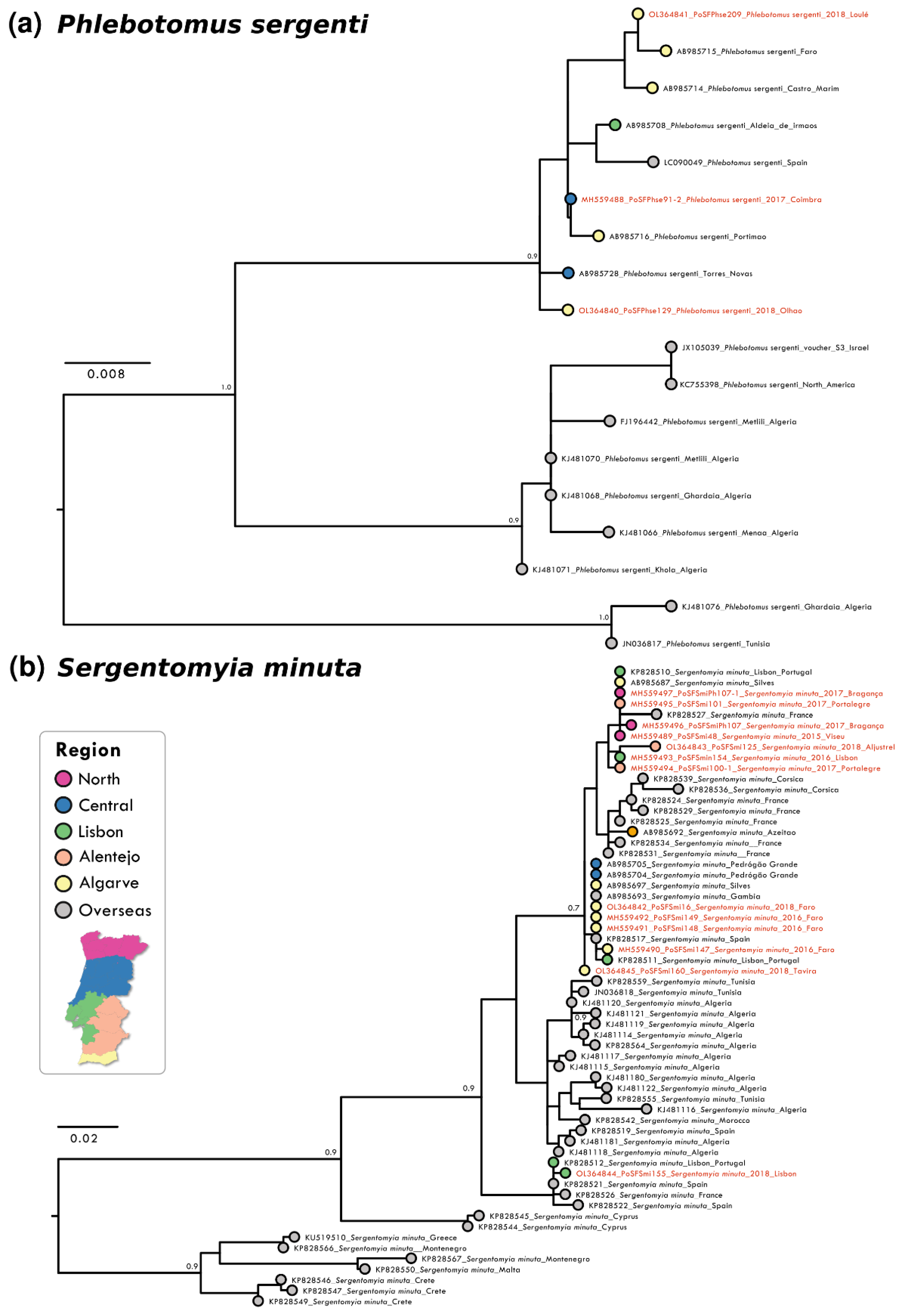
Analysis of partial DNA sequences from the COX1 gene obtained from sandflies collected from 2014 to 2018 showed that the number of polymorphic sites and parsimony informative sites was low for all species, reinforcing the high level of intra-specific conservation and the potential of the analysed gene region as bar coding. The highest conservation degree was observed in P. pernicious sequences with only six parsimony informative sites (p.i.s.) out of 452 nucleotides (nt) (totalling 1.32%). Nucleotide diversity per site was the lowest at π = 0.00040 for this species amongst the analyzed species’ sequences. This conservation was also confirmed in the resulting maximum likelihood (ML) phylogenetic reconstruction that did not show supported clusters (Figure 1b). P. ariasi sequence information (27 p.i.s. / 451 nt; π = 0.01275) indicated the presence of a genetically diverse population in the southernmost region of Portugal (Figure 1a). For P. sergenti a clear clustering of Iberian sequences was observed as well as two clusters with African/overseas populations (Figure 2a). S. minuta COX1 partial sequence analyses showed the highest number of p.i.s. (at 44 out of 388 nt, totally 11.34%; with π = 0.00295). Three well-supported clusters were obtained in the S. minuta ML tree (Figure 2b). The Portuguese sequences grouped together with European and African sequences, and two divergent clusters were formed with sequences from Cyprus and Crete, Greece, Malta, and Montenegro, respectively. G+C content was similar in all sequences ranging from 33.6% (P. perniciosus) to 38.0% (P. sergenti).
Figure 1.
Maximum likelihood (ML) phylogenetic trees of: (a) P. ariasi and (b) P. perniciosus were inferred using partial COX1 sequence with MEGA version 10 software. Distance matrices were calculated based on the Tamura 3-parameter model. Bootstrap values obtained from 500 replicate trees are shown for key nodes (>0.7). Scale bar at the bottom represents substitutions per site. GenBank accession number, place, and year of collection are indicated. Coloured bullets associated to geographical locations of each sequence are presented. The sequences related to this work are highlighted in red.
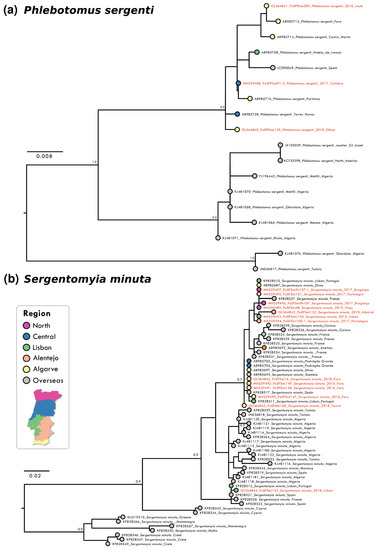
Figure 2.
Maximum likelihood (ML) phylogenetic trees of: (a) P. sergenti; (b) S. minuta. were inferred using partial COX1 sequence with MEGA version 10 software. Distance matrices were calculated based on the Tamura 3-parameter model (a) and Tamura-Nei model (b). Bootstrap values obtained from 500 replicate trees are shown for key nodes (>0.7). Scale bar at the bottom represents substitutions per site. GenBank accession number, place, and year of collection are indicated. Coloured bullets associated to geographical locations of each sequence are presented. The sequences related to this work are highlighted in red.
3.2. Distribution of Samples and Ecological Metadata
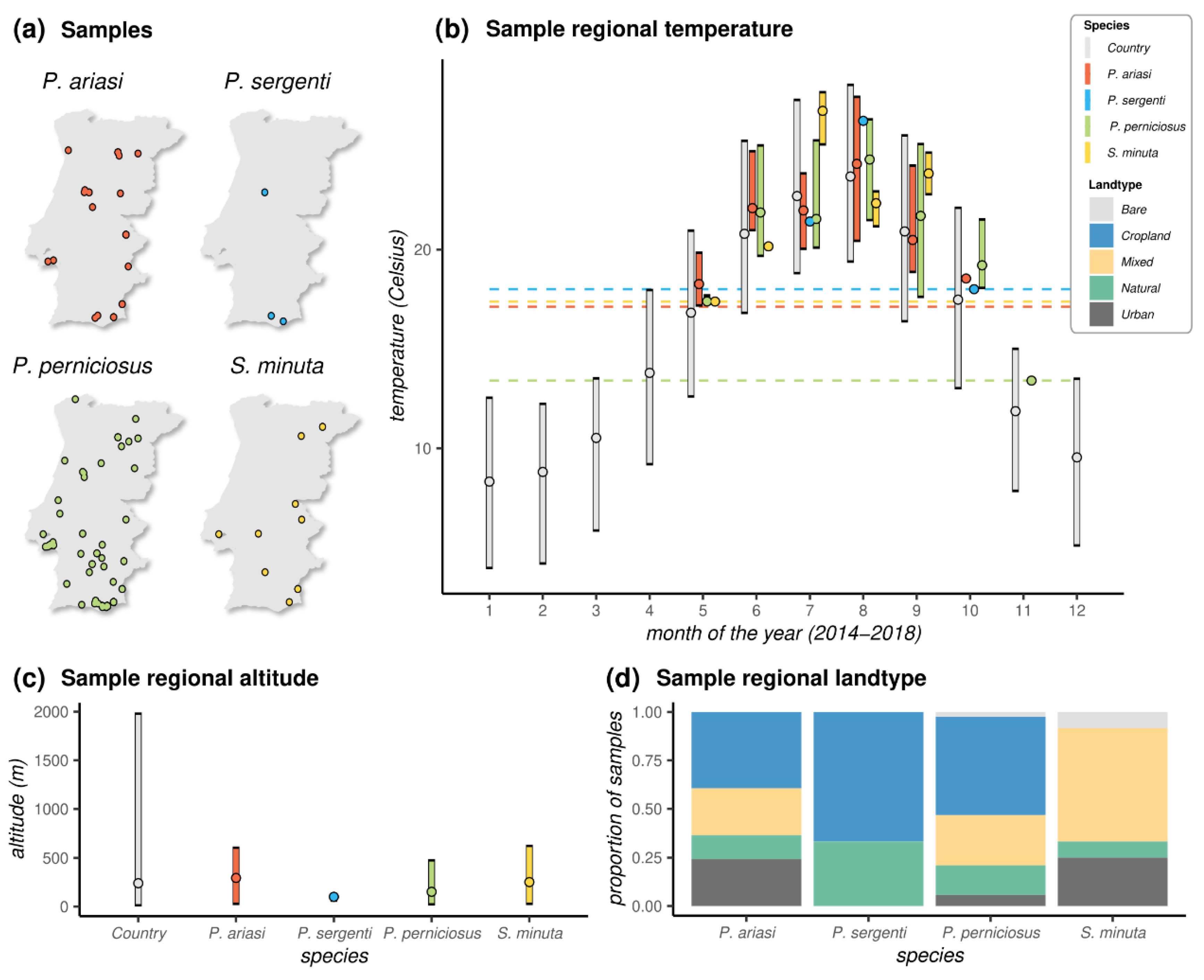
Most species, with the exception of P. sergenti for which only three samples existed, were found across the longitude and latitude ranges of Portugal (Figure 3a, Table S1). Together, the distribution of sandflies irrespective of species was virtually uniform across the territory. The southernmost site where adult sandflies were collected was in Olhão county in Faro (Algarve region, latitude 37.040103, P. perniciosus), and the northernmost site was in Mirandela county in Bragança (North region, latitude 41.557417, P. perniciosus). The westernmost site was in Loures county in Lisboa (Lisbon region, longitude -9.134300, P. perniciosus and S. minuta), and the easternmost site was in Vila Nova de Foz Côa in Guarda (Central region, longitude -7.141333, P. ariasi and P. perniciosus).
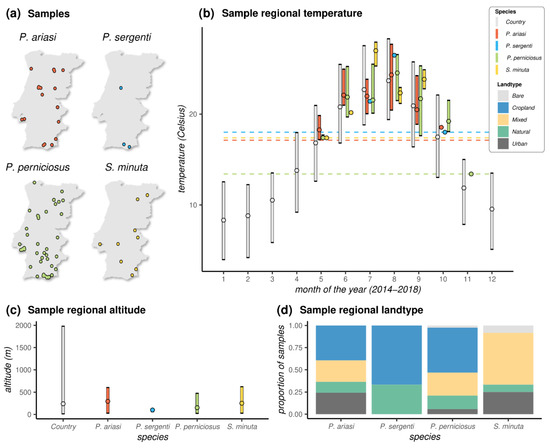
Figure 3.
Sample spatial, temperature, and land-type distributions: (a) Spatial location of each sample according to species; (b) distributions of observed local (sample) temperature (data are aggregated per month (1–12) over the observation period (2014–2018), and dashed horizontal lines mark the minimum temperature (Celsius, °C) observed per species (P. ariasi 17.12 °C; P. perniciosus 13.40 °C; P. sergenti 18.00 °C; S. minuta 17.38 °C); (c) distributions of observed local (sample) altitude; (d) proportion of samples (per species) according to local land-type classification (see Materials and Methods). Land types coloured according to legend on the right. (a–c) The “country” entry represents the observed values of the variables across Portugal. Points are the mean. Ranges are the 95% quantiles. For some species, when a simple sample was collected, a point is presented instead of a distribution. Data coloured per species according to legend on the right.
The longer range of seasonal adult sandflies was found for P. perniciosus, from 5 May in Faro (Algarve region) to 10 November in Santarém (Lisbon region) (Figure 3b). The seasonal detection range of the remaining species was from 16 May to 23 October for P. ariasi, 12 July to 9 October for P. sergenti, and 16 May to 11 September for S. minuta. These time windows were generally compatible with the months of the year presenting with temperatures in excess of 13 Celsius (°C). Indeed, with the exception of a single sample from P. perniciosus sampled in November when the mean temperature was 13.4 °C, all samples were found at mean monthly temperatures above 17 °C. Looking at the range of temperatures across the country over the months for the observation period (2014–2018), this 17 °C threshold mirrored the Portuguese summer months, including all months between May and October.
Across samples, the lowest altitude was at 5 m above sea level in Albufeira county in Faro (Algarve region, P. perniciosus), and the highest was at 770 m in Tabuaço county in Viseu (Central region, P. ariasi). Although a small country with a vast costal area, Portugal presents a variety of landscapes with a wide range of altitudes (Figure 3c). The distributions of sample altitudes across species did not appear to be different, although as expected due to detrimental effects of altitude on sandflies’ life cycle [42], the observed ranges were on the lower end of the country’s distribution. The proportion of samples collected in different land types presented some similarities among the three Phlebotomus species when compared to Sergentomya (Figure 3d). In particular, we observed a predominance of samples from locations characterized by cropland vegetation among the Phlebotomus species (~40% for P. ariasi, ~50% for P. perniciosus, and ~66% for P. sergenti). In contrast, there were no samples found in croplands for Sergentomyia, for which the most common land type was one characterized by a mix of natural and cropland vegetation (~58% of samples). Important for public health was the finding of both Phlebotomus and Sergentomyia species in land types classified as urban.
3.3. Blood Meals
A total of 37 screened females, the vast majority (97%), had fed only in humans. The exceptions were one P. perniciosus presenting a mixed bloodmeal for which it was not possible to determine the host source and one P. ariasi that had fed on a poultry (Gallus gallus).
4. Discussion
Patterns of vector-borne disease transmission are directly related to the vector’s population dynamics, geographic distribution, and genetic structure [21,22]. For sandflies and their pathogens, these links can depend, e.g., on climate variation affecting population dynamics [23], the landscape and land type affecting the spatial distribution [43], and the spatio-temporal presence of cryptic species or genetic variants (lineages) [44], which may vary in their susceptibility to the pathogen of interest [44], feeding behaviour including degrees of anthropophily and animal preference [45], female longevity [46], etc. In the European context, there is a lack of institutionalised sandfly surveillance, and control infrastructure for sandfly-borne pathogens is missing (e.g., human leishmaniasis and TOSV infections are not included in the list of diseases under EU surveillance) [21]. As such, exploring data from ongoing, potentially non-specific surveillance initiatives that may reveal factors dictating or associated with sandfly population dynamics, spatial distribution and genetic structure are key for our understanding of their possible impact on human and animal health.
Studies about the distribution of sandflies in Portugal are very scarce and mainly local [9,20,22,25,26,27,28]. Previous studies have shown that several species of sandflies are distributed across Portugal [26]. For P. perniciosus, our results confirm previous reports [13,26] finding that it is the most widespread species and that P. ariasi is typically the second, although in some regions of Portugal the latter species may be dominant [27]. S. minuta thrives in diverse habitats, but their low light attractiveness [22] may negatively bias the number of specimens collected in this study. Although it is known to be less frequent in the north [26], one of our samples had origin in a northern county, which means that more systematic studies using different collection methods need to be performed to clarify the real distribution and abundance. The least-represented species in our samples was P. sergenti, which according to other authors, is considered to have considerably low abundance in the country, with a possible exception of the southern regions of Algarve and Alentejo [28]. Sandflies have previously been found at varying altitudes, ranging from below sea level in areas surrounding the Dead Sea in Israel and Jordan to 2800 m above sea level in the Andes and Ethiopia [47]. In our dataset, sandflies were found at the lower end of the altitude variation found in Portugal, with the lowest observed altitude of 5 m and a highest of 770 m above sea level.
Besides climate and altitude, a third major determinant of sandfly spatial distribution is land type, which critically also determines the presence and abundance of vertebrates and thus the potential of sandfly species to participate in vector-borne pathogen transmission. In our dataset, we found that Portuguese sandflies were preferentially associated with cropland or mixed land (cropland and natural vegetation). There was nonetheless a reasonable proportion of samples found in urban areas, in particular for P. ariasi and S. minuta. Given the unspecific sampling nature of the dataset, it was not possible to discern if this observation was due to sampling being mostly from mosquito surveys performed in places with human presence.
Climate is a major determinant of the population dynamics of human and animal arthropod vectors, such as mosquitoes, ticks, and sandflies. For sandflies [48,49,50] and mosquitoes [51,52], temperature is the main determinant due to its effects on life-span and developmental rates. Importantly, variation exists in the response to climate between sandfly species and within species variants. For example, P. ariasi seemingly prefers wetter and milder conditions (e.g., forests), while P. perniciosus is more commonly associated with drier and warmer conditions (e.g., plains) [53]. Similarly, variants of P. sergenti from Morrocco appear less tolerant to low temperatures than those from Spain [54]. In general, seasonal variations in climate influence the intra-year, monthly abundance of sandflies. In Portugal, it has been previously reported that sandfly abundance/presence is associated with periods between April/May and October/November of each year [9,44,45]. Out of this period, it is generally agreed that a developmental bottleneck is introduced by low temperatures that may vary by species (e.g., 15 °C for P. perniciosus [55] and 16 °C for P. papatasi [56].
In the analyses of our sandfly samples, we observed intra-year temporal windows compatible with the aforementioned knowledge on the timing of Portuguese sandfly seasons and on the effect of temperature. Namely, we found that sandflies were only present in collections between May and October for a temperature threshold above 17 °C. The exception was a single sample of P. perniciosus from November with associated temperature of 13.4 °C. While thresholds as low as 13 °C have been described for some species (e.g., P. neglectus [57]), and considering the restricted and non-specific sampling of the presented dataset, it remains to be shown if any species of sandfly is commonly abundant in Portugal at such low temperatures typical of March and November.
Although we screened a total of six samples from henneries (Table S1), only one specimen of P. ariasi had fed on poultry, while the rest had fed on humans. It is not entirely surprising that specimens of P. ariasi, P. perniciosus, and P. sergenti were found engorged with human blood since they are recognised vectors of human pathogens in the Mediterranean Basin [44]. However, the fact that 97% of the bloodmeals were in humans is unexpected since sandflies are often reported to feed on a variety of vertebrates. Indeed, the feeding behaviour of P. ariasi, P. sergenti, and P. perniciosus has been reported to be similar and of opportunistic nature according to whatever vertebrate is of easier access, including humans, domesticated vertebrates, and birds [1,42,58]. On the other hand, sandflies from the genus Sergentomyia were previously believed to feed only on reptiles [59], although recent studies have shown that S. minuta also feeds on mammals, including humans [60]. All analysed engorged sandflies resulted from captures in mosquito surveys performed in places with human presence, and although some of them were rural environments with the presence of other animals, this is likely the reason why human bloodmeals dominated the dataset.
Our genetic analysis confirmed the previously reported utility of COX1 phylogenetic analysis as a tool in sandfly species identification [24]. This approach enabled species identification of all samples tested, due to the high level of intra-specific conservation observed for this gene region in all studied species. Despite the high level of conservation of the COX1 sequences analysed, two lineages were clearly observed in P. perniciosus, in agreement with previous reports [61,62]: the “typical lineage” with sequences found in Tunisia, Algeria, and Italy and the “Iberian lineage,” clustering all Portuguese with Spanish sequences. This clustering of Iberian sequences (including Portugal, Spain) is also observed in P. ariasi and P. sergenti, probably diverging from an ancient African population (Figure 1 and Figure 2). In S. minuta although the proximity between Iberian sequences was also seen, a diverse evolution history seems to be shaping the observed genetic structure with a well-defined “Eastern Mediterrenean” cluster and a closer relation between African and Iberian samples. Further studies, including more Phlebotomine sandfly specimens (from widespread collection sites over their geographical distribution) and using highly variable gene sequences at the intra-specific level, as the 3′ end of the COX1 gene [63,64], the cytochrome b (CytB) gene [65,66], or even complete mitogenome sequences, will be necessary to clarify the Portuguese genetic diversity and evolutionary history.
In this study, we summarized and analysed a series of Portuguese sandfly samples collected over specific and mosquito-focused surveillance between 2014 and 2018. For a generalized overview of the sandfly population, we explored sample genetic population structure in the context of local and global known genetic sequences and explored their geographical distribution, seasonal timing and associated temperature, altitude, land type, and bloodmeals. To our knowledge, this study is the first nationwide report of sandflies in Portugal.
Due to the use of different sampling methods of the presented sandfly dataset, this work presents a series of limitations. It is unlikely that a complete characterization of the existing genetic diversity of the four species included was achieved or that the full seasonal timing and spatial distribution was captured with our analyses. The non-specific sampling is also likely the reason why the vast majority of bloodmeal results was associated with humans, although sandflies are expected to feed on a vast diversity of vertebrate hosts. Most of blood meal analysis (68%, from 2014 to 2016) were from sandflies captures using CDC light traps for mosquito collection, mostly in habitats under surveillance for being reported by population as “mosquito annoyance/high mosquito bite rate” related and as such were inherently biased towards the proximity to human populations. This, however, did not hamper our ability to confirm ecological expectations and to replicate expected observations, such as the finding of sandflies across the geographical scale of Portugal, a genetic structure highly associated with small scale geographical clustering, the predominance of samples during the summer months when temperatures are above 17 °C, and the association with lower altitudes. Overall, this study reiterates the need for investment in sandfly-specific surveillance to adequately characterize the existing populations and their potential for local vector-borne pathogens and relevance for human and animal public health.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/zoonoticdis2010003/s1, Table S1: Sandflies analysed in this study.
Author Contributions
Conceptualization, L.Z.-Z., F.A. and J.L.; methodology, L.Z.-Z., F.A., H.C.O. and J.L.; validation, L.Z.-Z.; M.G. and J.L.; formal analysis, L.Z.-Z.; F.A., H.C.O.; M.G. and J.L.; writing—original draft preparation, L.Z.-Z.; writing—review and editing, L.Z.-Z., F.A., M.G., J.L. and M.J.A.; visualization, L.Z.-Z.; M.G. and J.L.; project administration, F.A. and M.J.A.; funding acquisition, F.A. and M.J.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by the Portuguese Foundation for Science and Technology (FCT) through the project: “Phleboviruses in Portugal—vectors, pathogenesis and co-infections” (PTDC/DTP-SAP/0859/2014). J.L. was supported by a research lectureship by the Zoology Department of the University of Oxford. M.G. was supported by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data used and presented in this study are either available in public repositories as described in the methods section or are made available in the Supplementary File with this article.
Acknowledgments
We are grateful to The Ministry of Health/National Institute of Health (INSA) under the National Vector Surveillance Network—REVIVE for supporting this research. We are also grateful to the REVIVE team for the sandflies collection nationwide.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Svobodová, M.; Sádlová, J.; Chang, K.P.; Volf, P. Short report: Distribution and feeding preference of the sand flies Phlebotomus sergenti and P. papatasi in a cutaneous leishmaniasis focus in Sanliurfa, Turkey. Am. J. Trop. Med. Hyg. 2003, 68, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Maia, C.; Campino, L. Leishmaniasis in Portugal in the beginning of the XXI century. Anais Do IHMT 2018, 13, 25–28. [Google Scholar] [CrossRef]
- Quinnell, R.; Courtenay, O. Transmission, reservoir hosts and control of zoonotic visceral leishmaniasis. Parasitology 2009, 136, 1915–1934. [Google Scholar] [CrossRef] [PubMed]
- WHO Expert Committee on the Control of the Leishmaniases & World Health Organization. Control of the Leishmaniases: Report of a Meeting of the WHO Expert Commitee on the Control of Leishmaniases, Geneva, 22–26 March 2010; World Health Organization: Geneva, Switzerland, 2010; Available online: https://apps.who.int/iris/handle/10665/44412 (accessed on 9 December 2021).
- Pennisi, M.G.; Cardoso, L.; Baneth, G.; Bourdeau, P.; Koutinas, A.; Miró, G.; Oliva, G.; Solano-Gallego, L. LeishVet update and recommendations on feline leishmaniosis. Parasites Vectors 2015, 8, 302. [Google Scholar] [CrossRef] [PubMed]
- Maia, C.; Dantas-Torres, F.; Campino, L. Parasite Biology: The Reservoir Hosts. In The Leishmaniases: Old Neglected Tropical Diseases; Bruschi, F., Gradoni, L., Eds.; Springer: Cham, Switzerland, 2018; pp. 79–106. [Google Scholar] [CrossRef]
- Spada, E.; Perego, R.; Vitale, F.; Bruno, F.; Castelli, G.; Tarantola, G.; Baggiani, L.; Magistrelli, S.; Proverbio, D. Feline Leishmania spp. infection in a Non-Endemic Area of Northern Italy. Animals 2020, 10, 817. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Parreira, R.; Cristóvão, J.M.; Vitale, F.; Bastien, P.; Campino, L.; Maia, C. Leishmania infantum strains from cats are similar in biological properties to canine and human strains. Vet. Parasitol. 2021, 298, 109531. [Google Scholar] [CrossRef]
- Pires, C.A. Phlebotomus of Portugal. I-Natural infestation of Phlebotomus ariasi Tonnoir, 1921 and Phlebotomus perniciosus Newstead, 1911, by Leishmania in the zoonotic focus of Arrábida (Portugal). Ann. Parasitol. Hum. Comp. 1984, 59, 521–524. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses. Virus Taxonomy Release: 2020 Release. Email Ratification March 2021 (MSL #36). Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 9 December 2021).
- Alkan, C.; Bichaud, L.; de Lamballerie, X.; Alten, B.; Gould, E.A.; Charrel, R.N. Sandfly-borne phleboviruses of Eurasia and Africa: Epidemiology, genetic diversity, geographic range, control measures. Antivir. Res. 2013, 100, 54–74. [Google Scholar] [CrossRef]
- Moriconi, M.; Rugna, G.; Calzolari, M.; Bellini, R.; Albieri, A.; Angelini, P.; Cagarelli, R.; Landini, M.P.; Charrel, R.N.; Varani, S. Phlebotomine sand fly-borne pathogens in the Mediterranean Basin: Human leishmaniasis and phlebovirus infections. PLOS Negl. Trop. Dis. 2017, 11, e0005660. [Google Scholar] [CrossRef]
- Amaro, F.; Zé-Zé, L.; Alves, M.J.; Börstler, J.; Clos, J.; Lorenzen, S.; Becker, S.C.; Schmidt-Chanasit, J.; Cadar, D. Co-circulation of a novel phlebovirus and Massilia virus in sandflies, Portugal. Virol. J. 2015, 12, 174. [Google Scholar] [CrossRef]
- Amaro, F.; Hanke, D.; Zé-Zé, L.; Alves, M.J.; Becker, S.C.; Höper, D. Genetic characterization of Arrabida virus, a novel phlebovirus isolated in South Portugal. Virus Res. 2016, 214, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Amaro, F.; Zé-Zé, L.; Lourenço, J.; Giovanetti, M.; Becker, S.C.; Alves, M.J. Phylogenetic Analysis of Massilia phlebovirus in Portugal. Viruses 2021, 13, 1412. [Google Scholar] [CrossRef] [PubMed]
- Amaro, F.; Zé-Zé, L.; Luz, M.T.; Alves, M.J. Toscana Virus: Ten Years of Diagnostics in Portugal. Acta Med. Port. 2021, 34, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Service, M. Phlebotomine sand-flies (Phlebotominae). In Medical Entomology for Students; Cambridge University Press: Cambridge, UK, 2008; pp. 93–102. [Google Scholar] [CrossRef]
- França, C. Phlebotomus papatasi (Scopoli) et fièvre à Pappataci au Portugal. Bull. Soc. Pathol. Éxotique 1913, 6, 123–124. [Google Scholar]
- França, C. Notes sur les espèces portugaises du genre Phlebotomus. Bull. Soc. Pathol. Éxotique 1918, 11, 730–733. [Google Scholar]
- Alves-Pires, C.; Afonso, M.O.; Janz, J.G.; Semião-Santos, S.J. The phlebotomine sand flies of Portugal. XII. The phlebotomine of Évora Leishmaniasis focus (1999–2000). Acta Parasitológica Port. 2004, 11, 41–45. [Google Scholar]
- European Centre for Disease Prevention and Control. Phlebotomine sand flies—Factsheet for Experts. 2015. Available online: https://www.ecdc.europa.eu/en/disease-vectors/facts/phlebotomine-sand-flies (accessed on 28 December 2021).
- Alten, B.; Maia, C.; Afonso, M.O.; Campino, L.; Jiménez, M.; González, E.; Molina, R.; Bañuls, A.L.; Prudhomme, J.; Vergnes, B.; et al. Seasonal dynamics of phlebotomine sand fly species proven vectors of Mediterranean leishmaniasis caused by Leishmania infantum. PLoS Negl. Trop. Dis. 2016, 10, e0004458. [Google Scholar] [CrossRef]
- Ballart, C.; Baron, S.; Alcover, M.M.; Portus, M.; Gallego, M. Distribution of phlebotomine sand flies (Diptera: Psychodidae) in Andorra: First finding of P. perniciosus and wide distribution of P. ariasi. Acta Trop. 2012, 122, 155–159. [Google Scholar] [CrossRef]
- Maia, C.; Parreira, R.; Cristóvão, J.M.; Afonso, M.O.; Campino, L. Exploring the utility of phylogenetic analysis of cytochrome oxidase gene subunit I as a complementary tool to classical taxonomical identification of phlebotomine sand fly species (Diptera, Psychodidae) from southern Europe. Acta. Trop. 2015, 144, 1–8. [Google Scholar] [CrossRef]
- Pereira, S.; Pita-Pereira, D.; Araujo-Pereira, T.; Britto, C.; Costa-Rego, T.; Ferrolho, J.; Vilhena, M.; Rangel, E.F.; Vilela, M.L.; Afonso, M.O. First molecular detection of Leishmania infantum in Sergentomyia minuta (Diptera, Psychodidae) in Alentejo, southern Portugal. Acta Trop. 2017, 174, 45–48. [Google Scholar] [CrossRef]
- Pires, C. Os Flebótomos (Diptera, Psychodidae) dos Focos Zoonóticos de Leishmanioses em Portugal. Ph.D. Thesis, Universidade Nova de Lisboa, Instituto de Higiene e Medicina Tropical, Lisboa, Portugal, 2000. (In Portuguese). [Google Scholar]
- Alves-Pires, C.; Ribeiro, H. The phlebotomine sandflies of Portugal. V—Observations of the ecology of the vectors of leishmaniasis in the Alto-Douro region. Parasitologia 1991, 33, 63–68. [Google Scholar]
- Afonso, M.O.; Campino, L.; Cortes, S.; Alves-Pires, C. The phlebotomine sandflies of Portugal. XIII--Occurrence of Phlebotomus sergenti Parrot, 1917 in the Arrabida leishmaniasis focus. Parasite 2005, 12, 69–72. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Osório, H.C.; Zé-Zé, L.; Amaro, F.; Alves, M.J. Mosquito surveillance for prevention and control of emerging mosquito-borne diseases in Portugal—2008–2014. Int. J. Environ. Res. Public Health 2014, 11, 11583–11596. [Google Scholar] [CrossRef] [PubMed]
- Topographic-map.com. Available online: https://pt-br.topographic-map.com/maps/zhkm/ (accessed on 29 December 2021).
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial Cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Osório, H.C.; Zé-Zé, L.; Neto, M.; Silva, S.; Marques, F.; Silva, A.S.; Alves, M.J. Detection of the invasive mosquito species Aedes (Stegomya) albopictus (Diptera: Culicidae) in Portugal. Int. J. Environ. Res. Public Health 2018, 15, E820. [Google Scholar] [CrossRef]
- Molaei, G.; Andreadis, T.G. Identification of avian- and mammalian-derived bloodmeals in Aedes vexans and Culiseta melanura (Diptera: Culicidae) and its implication for West Nile virus transmission in Connecticut, U.S.A. J. Med. Entomol. 2006, 43, 1088–1093. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Land Cover Classification Gridded Maps from 1992 to Present Derived from Satellite Observations. Available online: https://cds.climate.copernicus.eu/cdsapp#!/dataset/satellite-land-cover?tab=overview (accessed on 15 December 2021).
- Essential Climate Variables for Assessment of Climate Variability from 1979 to Present. Available online: https://cds.climate.copernicus.eu/cdsapp#!/dataset/ecv-for-climate-change (accessed on 14 December 2021).
- Lourenço, J.; Barros, S.C.; Zé-Zé, L.; Damineli, D.C.S.; Giovanetti, M.; Osório, H.C.; Amaro, F.; Henriques, A.M.; Ramos, F.; Luís, T.; et al. West Nile virus transmission potential in Portugal. Comm. Biol. 2022, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Depaquit, J.; Hadj-Henni, L.; Bounamous, A.; Strutz, S.; Boussaa, S.; Morillas-Marquez, F.; Pesson, B.; Gállego, M.; Delécolle, J.C.; Afonso, M.O.; et al. Mitochondrial DNA Intraspecific Variability in Sergentomyia minuta (Diptera: Psychodidae). J. Med. Entomol. 2015, 52, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Ballart, C.; Guerrero, I.; Castells, X.; Barón, S.; Castillejo, S.; Alcover, M.M.; Portús, M.; Gállego, M. Importance of individual analysis of environmental and climatic factors affecting the density of Leishmania vectors living in the same geographical area: The example of Phlebotomus ariasi and P. perniciosus in northeast Spain. Geospat. Health 2014, 8, 389–403. [Google Scholar] [CrossRef] [PubMed]
- McCoy, K.D. The population genetic structure of vectors and our understanding of disease epidemiology. Parasite 2008, 15, 444–448. [Google Scholar] [CrossRef] [PubMed]
- De Colmenares, M.; Portús, M.; Botet, J.; Dobaño, C.; Gállego, M.; Wolff, M.; Seguí, G. Identification of blood meals of Phlebotomus perniciosus (Diptera: Psychodidae) in Spain by a competitive enzyme-linked immunosorbent assay biotin/avidin method. J. Med. Entomol. 1995, 32, 229–233. [Google Scholar] [CrossRef]
- Scarpassa, V.M.; Figueiredo, A.S.; Alencar, R.B. Genetic diversity and population structure in the Leishmania guyanensis vector Lutzomyia anduzei (Diptera, Psychodidae) from the Brazilian Amazon. Infect. Genet. Evol. 2015, 31, 312–320. [Google Scholar] [CrossRef]
- Killick-Kendrick, R.; Killick-Kendrick, M. Biology of sand fly vectors of Mediterranean canine leishmaniasis. In Canine Leishmaniasis: An Update. Proceedings of the International Canine Leishmaniasis Forum, Barcelona, Spain, 20–24 May 1999; Hoechst Roussel Vet: Wiesbaden, Germany, 1999; pp. 26–31. [Google Scholar]
- Benkova, I.; Volf, P. Effect of temperature on metabolism of Phlebotomus papatasi (Diptera: Psychodidae). J. Med. Entomol. 2007, 44, 150–154. [Google Scholar] [CrossRef]
- Trájer, A.J.; Bede-Fazekas, A.; Hufnagel, l.; Horvath, L.; Bovbos, J. The effect of climate change on the potential distribution of the European Phlebotomus species. Appl. Ecol. Env. Res. 2013, 11, 189–208. [Google Scholar] [CrossRef]
- Risueño, J.; Muñoz, C.; Pérez-Cutillas, P.; Goyena, E.; Gonzálvez, M.; Ortuño, M.; Bernal, L.J.; Ortiz, J.; Alten, B.; Berriatua, E. Understanding Phlebotomus perniciosus abundance in south-east Spain: Assessing the role of environmental and anthropic factors. Parasit. Vectors 2017, 10, 189. [Google Scholar] [CrossRef]
- Obolski, U.; Perez, P.N.; Villabona-Arenas, C.J.; Thézé, J.; Faria, N.R.; Lourenço, J. MVSE: An R-package that estimates a climate-driven mosquito-borne viral suitability index. Methods Ecol. Evol. 2019, 10, 1357–1370. [Google Scholar] [CrossRef] [PubMed]
- Bellone, R.; Failloux, A.B. The Role of Temperature in Shaping Mosquito-Borne Viruses Transmission. Front. Microbiol. 2020, 11, 584846. [Google Scholar] [CrossRef] [PubMed]
- Chamaillé, L.; Tran, A.; Meunier, A.; Bourdoiseau, G.; Ready, P.; Dedet, J.P. Environmental risk mapping of canine leishmaniasis in France. Parasit. Vectors 2010, 3, 31. [Google Scholar] [CrossRef] [PubMed]
- Merino-Espinosa, G.; Corpas-Lopez, V.; Callejon-Fernandez, R.; Porcel-Rodriguez, L.; Diaz-Saez, V.; Gallego, M.; Ballart, C.; Molina, R.; Jiménez, M.; Morillas-Márquez, F.; et al. Differential ecological traits of two Phlebotomus sergenti mitochondrial lineages in southwestern Europe and their epidemiological implications. Trop. Med. Int. Health 2016, 21, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Casimiro, E.; Calheiros, J.; Santos, F.D.; Kovats, S. National assessment of human health effects of climate change in Portugal: Approach and key findings. Environ. Health Perspect. 2006, 114, 1950–1956. [Google Scholar] [CrossRef] [PubMed]
- Colacicco-Mayhugh, M.G.; Masuoka, P.M.; Grieco, J.P. Ecological niche model of Phlebotomus alexandri and P. papatasi (Diptera: Psychodidae) in the Middle East. Int. J. Health Geogr. 2010, 9, 2. [Google Scholar] [CrossRef]
- Trájer, A.J.; Sebestyén, V. The changing distribution of Leishmania infantum Nicolle, 1908 and its Mediterranean sandfly vectors in the last 140 kys. Sci. Rep. 2019, 9, 11820. [Google Scholar] [CrossRef]
- Killick-Kendrick, R.; Rioux, J.A.; Bailly, M.; Guy, M.W.; Wilkes, T.J.; Guy, F.M.; Davidson, I.; Knechtli, R.; Ward, R.D.; Guilvard, E.; et al. Ecology of leishmaniasis in the south of France. 20. Dispersal of Phlebotomus ariasi Tonnoir, 1921 as a factor in the spread of visceral leishmaniasis in the Cévennes. Ann. Parasitol. Hum. Comp. 1984, 59, 555–572. [Google Scholar] [CrossRef]
- Charrel, R.N.; Izri, A.; Temmam, S.; de Lamballerie, X.; Parola, P. Toscana virus RNA in Sergentomyia minuta flies. Emer. Infect. Dis. 2006, 12, 1299–1300. [Google Scholar] [CrossRef]
- González, E.; Molina, R.; Aldea, I.; Iriso, A.; Tello, A.; Jiménez, M. Leishmania sp. detection and blood-feeding behaviour of Sergentomyia minuta collected in the human leishmaniasis focus of southwestern Madrid, Spain (2012–2017). Transbound. Emerg. Dis. 2020, 67, 1393–1400. [Google Scholar] [CrossRef]
- Aransay, A.; Ready, P.; Morillas-Marquez, F. Population differentiation of Phlebotomus perniciosus in Spain following postglacial dispersal. Heredity 2003, 90, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Perrotey, S.; Mahamdallie, S.S.; Pesson, B.; Richardson, K.J.; Gállego, M.; Ready, P. Postglacial dispersal of Phlebotomus perniciosus into France. Parasite 2006, 12, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Arrivillaga, J.C.; Norris, D.E.; Feliciangeli, M.D.; Lanzaro, G.C. Phylogeography of the neotropical sand fly Lutzomyia longipalpis inferred from mitochondrial DNA sequences. Infect. Genet. Evol. 2002, 2, 83–95. [Google Scholar] [CrossRef]
- Scarpassa, V.M.; Alencar, R.B. Lutzomyia umbratilis, the Main Vector of Leishmania guyanensis, Represents a Novel Species Complex? PLoS ONE 2012, 7, e37341. [Google Scholar] [CrossRef]
- Pech-May, A.; Marina, C.F.; Vázquez-Domínguez, E.; Berzunza-Cruz, M.; Rebollar-Téllez, E.A.; Narváez-Zapata, J.A.; Moo-Llanes, D.; Ibáñez-Bernal, S.; Ramsey, J.M.; Becker, I. Genetic structure and divergence in populations of Lutzomyia cruciata, a phlebotomine sand fly (Diptera: Psychodidae) vector of Leishmania mexicana in southeastern Mexico. Infect. Genet. Evol. 2013, 16, 254–262. [Google Scholar] [CrossRef]
- Yamamoto, K.; Cáceres, A.G.; Gomez, E.A.; Mimori, T.; Iwata, H.; Korenaga, M.; Sakurai, T.; Katakura, K.; Hashiguchi, Y.; Kato, H. Genetic diversity of the mitochondrial cytochrome b gene in Lutzomyia spp., with special reference to Lutzomyia peruensis, a main vector of Leishmania (Viannia) peruviana in the Peruvian Andes. Acta Trop. 2013, 126, 156–163. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).