Genetic Diversity, Antimicrobial Resistance and Survival upon Manure Storage of Campylobacter jejuni Isolated from Dairy Cattle Farms in the Cantabric Coast of Spain
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Media and Growth Conditions
2.2. Sample Collection, Processing and C. jejuni Strains Isolation
2.3. Molecular Methods for Species Identification and Strain Characterization
2.4. Testing of Susceptibility to Antimicrobial Agents
2.5. Decline of C. jejuni in Manure
2.6. Decline of C. jejuni in Pasture Crops
2.7. Analysis of Physical and Chemical Parameters of the Livestock Wastes
3. Results
3.1. Isolation and Characterization of C. jejuni Strains
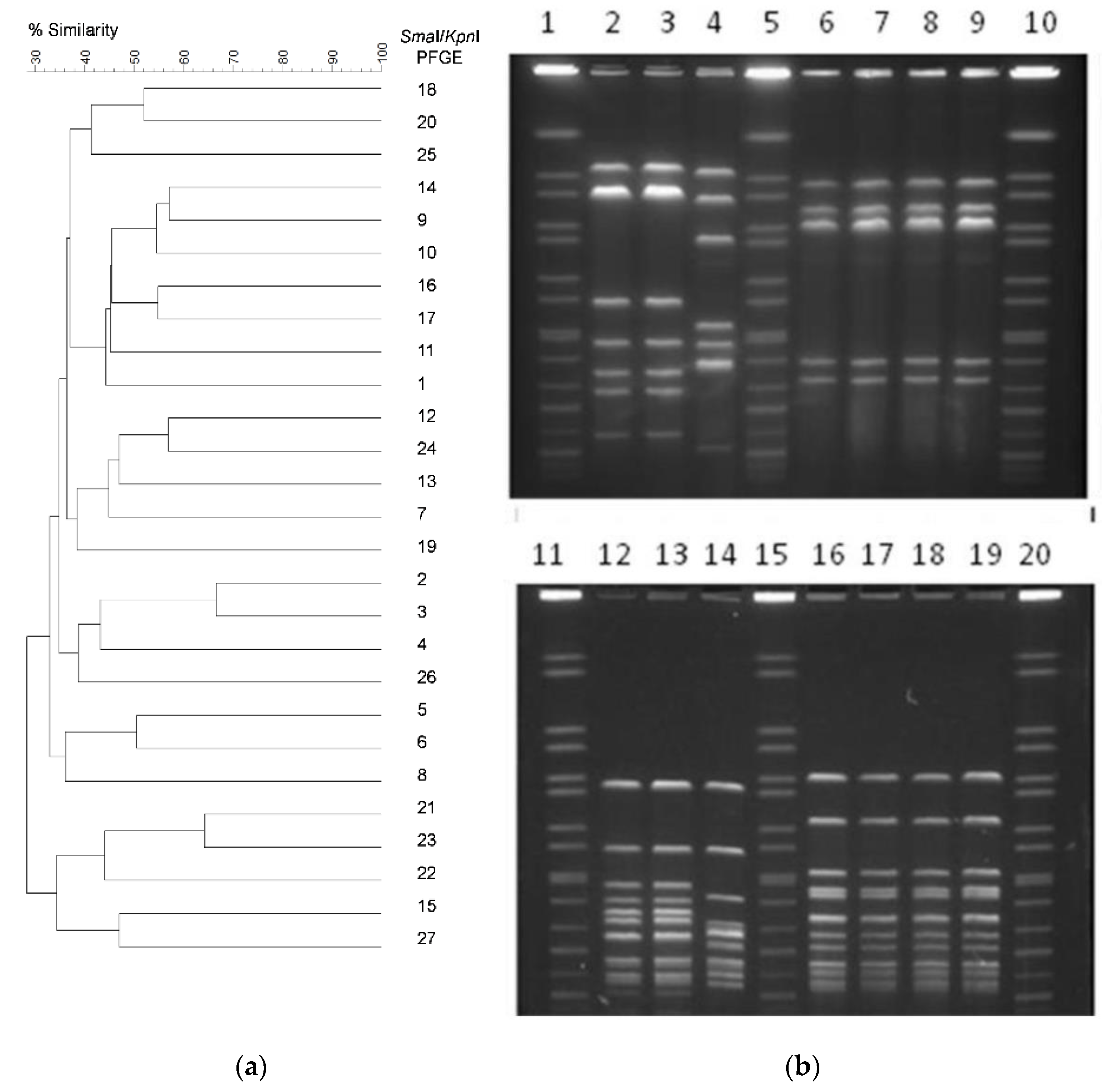
3.2. Genetic Diversity of C. jejuni Isolates
3.3. Antimicrobial Susceptibility Testing
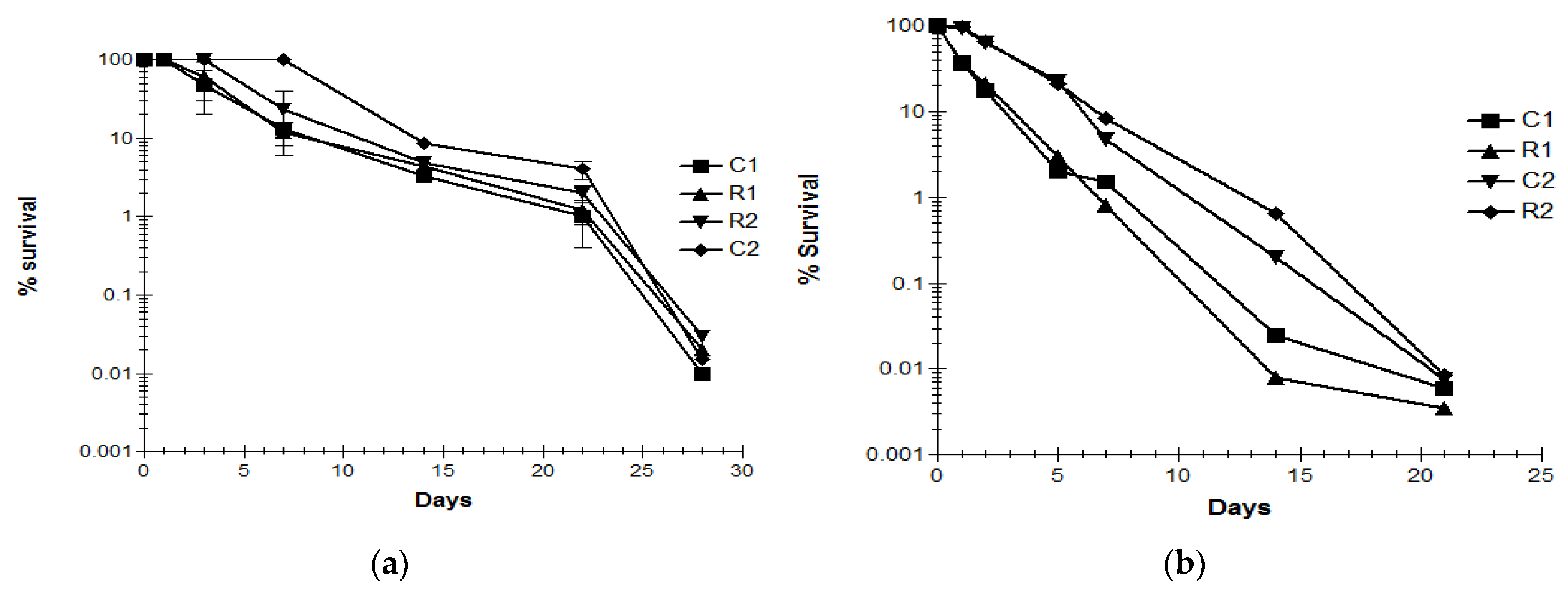
3.4. Decline of C. jejuni Strains in Livestock Waste
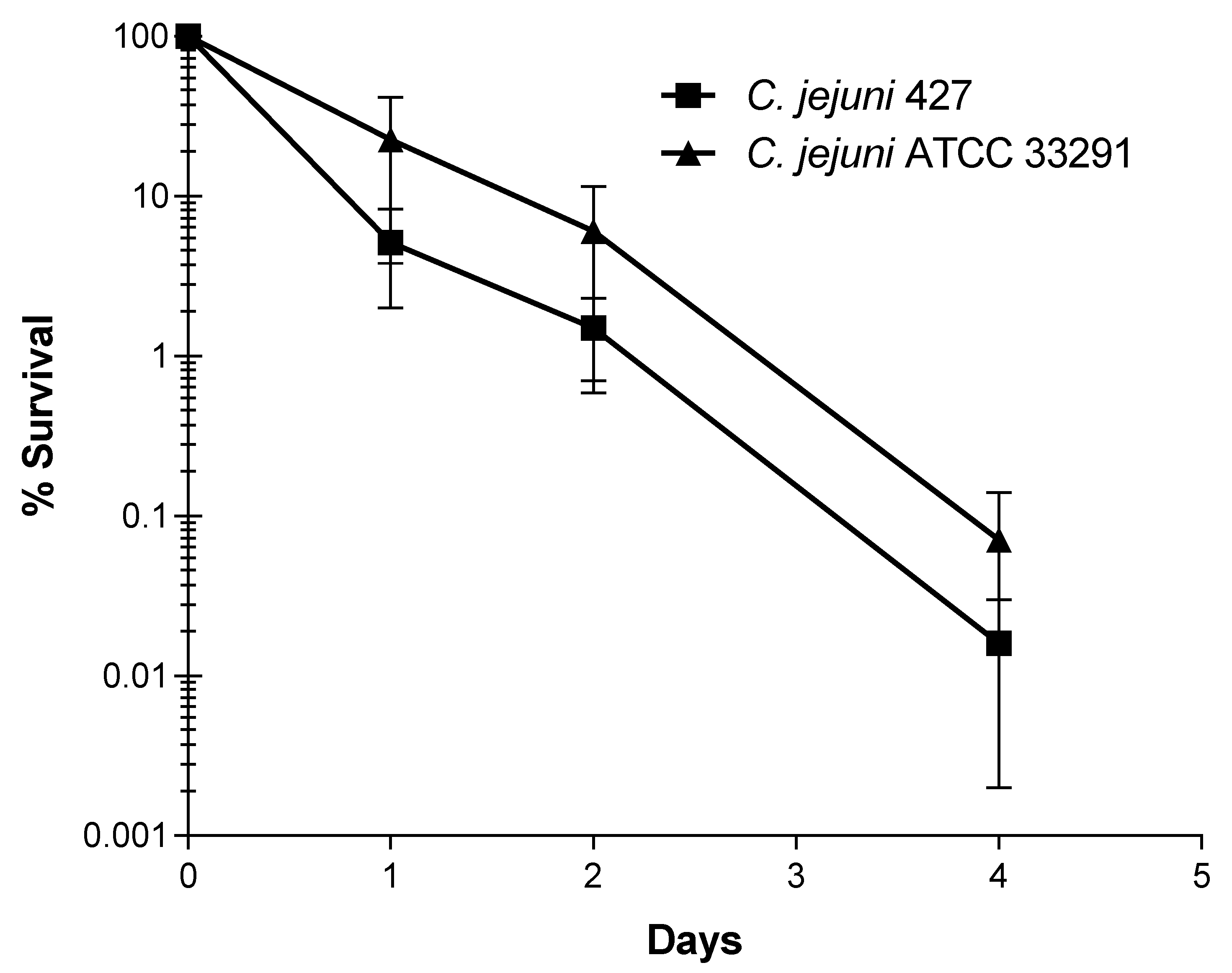
3.5. Decline of C. jejuni Strains upon Application of Infected Manure to Crops
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EFSA. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar] [CrossRef]
- Ragimbeau, C.; Schneider, F.; Losch, S.; Even, J.; Mossong, J. Multilocus Sequence Typing, Pulsed-Field Gel Electrophoresis, and Fla Short Variable Region Typing of Clonal Complexes of Campylobacter jejuni Strains of Human, Bovine, and Poultry Origins in Luxembourg. Appl. Environ. Microbiol. 2008, 74, 7715–7722. [Google Scholar] [CrossRef] [PubMed]
- Stanley, K.; Jones, K. Cattle and Sheep Farms as Reservoirs of Campylobacter. J. Appl. Microbiol. 2003, 94 (Suppl. S1), 104S–113S. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.R.; Roberts, R.J.; Ribeiro, C.D.; Gardner, D.; Kembrey, D. A Milk-Borne Campylobacter Outbreak Following an Educational Farm Visit. Epidemiol. Infect. 1996, 117, 457–462. [Google Scholar] [CrossRef]
- Altekruse, S.F.; Stern, N.J.; Fields, P.I.; Swerdlow, D.L. Campylobacter jejuni—An Emerging Foodborne Pathogen. Emerg. Infect. Dis. 1999, 5, 28–35. [Google Scholar] [CrossRef]
- Karagiannis, I.; Sideroglou, T.; Gkolfinopoulou, K.; Tsouri, A.; Lampousaki, D.; Velonakis, E.N.; Scoulica, E.V.; Mellou, K.; Panagiotopoulos, T.; Bonovas, S. A Waterborne Campylobacter jejuni Outbreak on a Greek Island. Epidemiol. Infect. 2010, 138, 1726–1734. [Google Scholar] [CrossRef]
- Janssen, R.; Krogfelt, K.A.; Cawthraw, S.A.; van Pelt, W.; Wagenaar, J.A.; Owen, R.J. Host-Pathogen Interactions in Campylobacter Infections: The Host Perspective. Clin. Microbiol. Rev. 2008, 21, 505–518. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance Global Report on Surveillance; WHO: Geneva, Switzerland, 2017; Available online: https://www.who.int/publications/i/item/9789241564748 (accessed on 1 January 2021).
- Mourkas, E.; Florez-Cuadrado, D.; Pascoe, B.; Calland, J.K.; Bayliss, S.C.; Mageiros, L.; Méric, G.; Hitchings, M.D.; Quesada, A.; Porrero, C.; et al. Gene Pool Transmission of Multidrug Resistance among Campylobacter from Livestock, Sewage and Human Disease. Environ. Microbiol. 2019, 21, 4597–4613. [Google Scholar] [CrossRef]
- Whelan, M.V.X.; Ardill, L.; Koide, K.; Nakajima, C.; Suzuki, Y.; Simpson, J.C.; Cróinín, Ó.T. Acquisition of Fluoroquinolone Resistance Leads to Increased Biofilm Formation and Pathogenicity in Campylobacter jejuni. Sci. Rep. 2019, 9, 18216. [Google Scholar] [CrossRef]
- Andrzejewska, M.; Szczepańska, B.; Śpica, D.; Klawe, J.J. Prevalence, Virulence, and Antimicrobial Resistance of Campylobacter spp. in Raw Milk, Beef, and Pork Meat in Northern Poland. Foods 2019, 8, 420. [Google Scholar] [CrossRef]
- Di Giannatale, E.; Calistri, P.; Di Donato, G.; Decastelli, L.; Goffredo, E.; Adriano, D.; Mancini, M.E.; Galleggiante, A.; Neri, D.; Antoci, S.; et al. Thermotolerant Campylobacter spp. in Chicken and Bovine Meat in Italy: Prevalence, Level of Contamination and Molecular Characterization of Isolates. PLoS ONE 2019, 14, e0225957. [Google Scholar] [CrossRef] [PubMed]
- Marotta, F.; Garofolo, G.; Di Marcantonio, L.; Di Serafino, G.; Neri, D.; Romantini, R.; Sacchini, L.; Alessiani, A.; Di Donato, G.; Nuvoloni, R.; et al. Antimicrobial Resistance Genotypes and Phenotypes of Campylobacter jejuni Isolated in Italy from Humans, Birds from Wild and Urban Habitats, and Poultry. PLoS ONE 2019, 14, e0223804. [Google Scholar] [CrossRef] [PubMed]
- Picó, Y.; Andreu, V. Fluoroquinolones in Soil--Risks and Challenges. Anal. Bioanal. Chem. 2007, 387, 1287–1299. [Google Scholar] [CrossRef]
- Lillenberg, M.; Yurchenko, S.; Kipper, K.; Herodes, K.; Pihl, V.; Lõhmus, R.; Ivask, M.; Kuu, A.; Kutti, S.; Litvin, S.V.; et al. Presence of Fluoroquinolones and Sulfonamides in Urban Sewage Sludge and Their Degradation as a Result of Composting. Int. J. Environ. Sci. Technol. 2010, 7, 307–312. [Google Scholar] [CrossRef]
- Ozawa, M.; Hiki, M.; Kawanishi, M.; Abo, H.; Kojima, A.; Asai, T.; Hamamoto, S. Molecular Typing of Fluoroquinolone-Resistant Campylobacter jejuni Isolated from Broilers in Japan Using Multilocus Sequence Typing and Pulsed-Field Gel Electrophoresis. Foodborne Pathog. Dis. 2016, 13, 1–7. [Google Scholar] [CrossRef]
- Guerra, B.; Fischer, J.; Helmuth, R. An Emerging Public Health Problem: Acquired Carbapenemase-Producing Microorganisms Are Present in Food-Producing Animals, Their Environment, Companion Animals and Wild Birds. Vet. Microbiol. 2014, 171, 290–297. [Google Scholar] [CrossRef]
- Castanon, J.I.R. History of the Use of Antibiotic as Growth Promoters in European Poultry Feeds. Poult. Sci. 2007, 86, 2466–2471. [Google Scholar] [CrossRef]
- Van Boven, M.; Veldman, K.T.; de Jong, M.C.M.; Mevius, D.J. Rapid Selection of Quinolone Resistance in Campylobacter jejuni but Not in Escherichia coli in Individually Housed Broilers. J. Antimicrob. Chemother. 2003, 52, 719–723. [Google Scholar] [CrossRef]
- Luo, N.; Pereira, S.; Sahin, O.; Lin, J.; Huang, S.; Michel, L.; Zhang, Q. Enhanced in Vivo Fitness of Fluoroquinolone-Resistant Campylobacter jejuni in the Absence of Antibiotic Selection Pressure. Proc. Natl. Acad. Sci. USA 2005, 102, 541–546. [Google Scholar] [CrossRef]
- Asakura, H.; Taguchi, M.; Ekawa, T.; Yamamoto, S.; Igimi, S. Continued Widespread Dissemination and Increased Poultry Host Fitness of Campylobacter jejuni ST-4526 and ST-4253 in Japan. J. Appl. Microbiol. 2013, 114, 1529–1538. [Google Scholar] [CrossRef]
- Haruna, M.; Sasaki, Y.; Murakami, M.; Ikeda, A.; Kusukawa, M.; Tsujiyama, Y.; Ito, K.; Asai, T.; Yamada, Y. Prevalence and Antimicrobial Susceptibility of Campylobacter in Broiler Flocks in Japan. Zoonoses Public Health 2012, 59, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Harada, K.; Ishihara, K.; Kojima, A.; Sameshima, T.; Tamura, Y.; Takahashi, T. Association of Antimicrobial Resistance in Campylobacter Isolated from Food-Producing Animals with Antimicrobial Use on Farms. Jpn. J. Infect. Dis. 2007, 60, 290–294. [Google Scholar] [PubMed]
- Allos, B.M. Campylobacter jejuni Infections: Update on Emerging Issues and Trends. Clin. Infect. Dis. 2001, 32, 1201–1206. [Google Scholar] [CrossRef]
- Guan, T.; Holley, R. Pathogen Survival in Swine Manure Environments and Transmission of Human Enteric Illness—A review. J. Environ. Qual. 2003, 32, 383–392. [Google Scholar] [CrossRef]
- Kashoma, I.P.; Kassem, I.I.; Kumar, A.; Kessy, B.M.; Gebreyes, W.; Kazwala, R.R.; Rajashekara, G. Antimicrobial Resistance and Genotypic Diversity of Campylobacter Isolated from Pigs, Dairy, and Beef Cattle in Tanzania. Front. Microbiol. 2015, 6, 1240. [Google Scholar] [CrossRef]
- Hutchison, M.L.; Walters, L.D.; Avery, S.M.; Moore, A. Decline of Zoonotic Agents in Livestock Waste and Bedding Heaps. J. Appl. Microbiol. 2005, 99, 354–362. [Google Scholar] [CrossRef]
- ISO 10272-1 ISO 10272-1:2006; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for Detection and Enumeration of Campylobacter spp.—Part 1: Detection Method. ISO: Geneva, Switzerland, 2006. Available online: https://www.iso.org/standard/37091.html (accessed on 11 February 2020).
- Linton, D.; Lawson, A.J.; Owen, R.J.; Stanley, J. PCR Detection, Identification to Species Level, and Fingerprinting of Campylobacter jejuni and Campylobacter coli Direct from Diarrheic Samples. J. Clin. Microbiol. 1997, 35, 2568–2572. [Google Scholar] [CrossRef] [PubMed]
- Ribot, E.M.; Fitzgerald, C.; Kubota, K.; Swaminathan, B.; Barrett, T.J. Rapid Pulsed-Field Gel Electrophoresis Protocol for Subtyping of Campylobacter jejuni. J. Clin. Microbiol. 2001, 39, 1889–1894. [Google Scholar] [CrossRef]
- Hunter, S.B.; Vauterin, P.; Lambert-Fair, M.A.; Van Duyne, M.S.; Kubota, K.; Graves, L.; Wrigley, D.; Barrett, T.; Ribot, E. Establishment of a Universal Size Standard Strain for Use with the Pulsenet Standardized Pulsed-Field Gel Electrophoresis Protocols: Converting the National Databases to the New Size Standard. J. Clin. Microbiol. 2005, 43, 1045–1050. [Google Scholar] [CrossRef]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting Chromosomal DNA Restriction Patterns Produced by Pulsed- Field Gel Electrophoresis: Criteria for Bacterial Strain Typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef]
- EUCAST European Committee on Antimicrobial Susceptibility Testing. Antimicrobial Susceptibility Testing EUCAST Disk Diffusion Methodology. 2022. Available online: https://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology/ (accessed on 1 January 2021).
- EUCAST European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. 2022. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 1 January 2021).
- Varsaki, A.; Murphy, C.; Barczynska, A.; Jordan, K.; Carroll, C. The Acid Adaptive Tolerance Response in Campylobacter jejuni Induces a Global Response, as Suggested by Proteomics and Microarrays. Microb. Biotechnol. 2015, 8, 974–988. [Google Scholar] [CrossRef] [PubMed]
- MAPA Ministry of Agriculture of Spain. Métodos Oficiales de Análisis III; Ministerio de Agricultura, Pesca y Alimentación: Madrid, Spain, 1994. Available online: https://www.mapa.gob.es/es/ (accessed on 1 January 2021).
- Olsen, S.R.; Watanabe, F.S.; Cosper, H.R.; Larson, W.E.; Nelson, L.B. Residual Phosphorus Availability in Long-Time Rotations on Calcareous Soils. Soil Sci. 1954, 78, 141–151. [Google Scholar] [CrossRef]
- Cook, K.L.; Netthisinghe, A.M.P.; Gilfillen, R.A. Detection of Pathogens, Indicators, and Antibiotic Resistance Genes after Land Application of Poultry Litter. J. Environ. Qual. 2014, 43, 1546–1558. [Google Scholar] [CrossRef]
- Rollins, D.M.; Colwell, R.R. Viable but Nonculturable Stage of Campylobacter jejuni and Its Role in Survival in the Natural Aquatic Environment. Appl. Environ. Microbiol. 1986, 52, 531–538. [Google Scholar] [CrossRef]
- Moran, A.P.; Upton, M.E. Factors Affecting Production of Coccoid Forms by Campylobacter jejuni on Solid Media during Incubation. J. Appl. Bacteriol. 1987, 62, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.L.; Bolster, C.H. Survival of Campylobacter jejuni and Escherichia coli in Groundwater during Prolonged Starvation at Low Temperatures. J. Appl. Microbiol. 2007, 103, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Yan, T.; Vu, H.; Hansen, D.L.; Hicks, R.E.; Sadowsky, M.J. Factors Controlling Long-Term Survival and Growth of Naturalized Escherichia coli Populations in Temperate Field Soils. Microbes Environ. 2010, 25, 8–14. [Google Scholar] [CrossRef]
- Strawn, L.K.; Fortes, E.D.; Bihn, E.A.; Nightingale, K.K.; Gröhn, Y.T.; Worobo, R.W.; Wiedmann, M.; Bergholz, P.W. Landscape and Meteorological Factors Affecting Prevalence of Three Food-Borne Pathogens in Fruit and Vegetable Farms. Appl. Environ. Microbiol. 2013, 79, 588–600. [Google Scholar] [CrossRef]
- Holley, R.; Walkty, J.; Blank, G.; Tenuta, M.; Ominski, K.; Krause, D.; Ng, L.-K. Examination of Salmonella and Escherichia coli Translocation from Hog Manure to Forage, Soil, and Cattle Grazed on the Hog Manure-Treated Pasture. J. Environ. Qual. 2008, 37, 2083–2092. [Google Scholar] [CrossRef]
- Ortiz, S.; López, V.; Villatoro, D.; López, P.; Dávila, J.C.; Martínez-Suárez, J.V. A 3-Year Surveillance of the Genetic Diversity and Persistence of Listeria monocytogenes in an Iberian Pig Slaughterhouse and Processing Plant. Foodborne Pathog. Dis. 2010, 7, 1177–1184. [Google Scholar] [CrossRef]
- Gu, W.; Siletzky, R.M.; Wright, S.; Islam, M.; Kathariou, S. Antimicrobial Susceptibility Profiles and Strain Type Diversity of Campylobacter jejuni Isolates from Turkeys in Eastern North Carolina. Appl. Environ. Microbiol. 2009, 75, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Griekspoor, P.; Engvall, E.O.; Olsen, B.; Waldenström, J. Multilocus Sequence Typing of Campylobacter jejuni from Broilers. Vet. Microbiol. 2010, 140, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Pergola, S.; Franciosini, M.P.; Comitini, F.; Ciani, M.; De Luca, S.; Bellucci, S.; Menchetti, L.; Casagrande Proietti, P. Genetic Diversity and Antimicrobial Resistance Profiles of Campylobacter coli and Campylobacter jejuni Isolated from Broiler Chicken in Farms and at Time of Slaughter in Central Italy. J. Appl. Microbiol. 2017, 122, 1348–1356. [Google Scholar] [CrossRef] [PubMed]
- Oporto, B.; Juste, R.A.; López-Portolés, J.A.; Hurtado, A. Genetic Diversity among Campylobacter jejuni Isolates from Healthy Livestock and Their Links to Human Isolates in Spain. Zoonoses Public Health 2011, 58, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gu, Y.; He, L.; Ran, L.; Xia, S.; Han, X.; Li, H.; Zhou, H.; Cui, Z.; Zhang, J. Molecular Typing and Antimicrobial Susceptibility Profiles of Campylobacter jejuni Isolates from North China. J. Med. Microbiol. 2010, 59, 1171–1177. [Google Scholar] [CrossRef]
- Noström, M.; Hofshagen, M.; Stavnes, T.; Schau, J.; Lassen, J.; Kruse, H. Antimicrobial Resistance in Campylobacter jejuni from Humans and Broilers in Norway. Epidemiol. Infect. 2006, 134, 127–130. [Google Scholar] [CrossRef]
- Van Looveren, M.; Daube, G.; De Zutter, L.; Dumont, J.M.; Lammens, C.; Wijdooghe, M.; Vandamme, P.; Jouret, M.; Cornelis, M.; Goossens, H. Antimicrobial Susceptibilities of Campylobacter Strains Isolated from Food Animals in Belgium. J. Antimicrob. Chemother. 2001, 48, 235–240. [Google Scholar] [CrossRef]
- Sáenz, Y.; Zarazaga, M.; Lantero, M.; Gastañares, M.J.; Baquero, F.; Torres, C. Antibiotic Resistance in Campylobacter Strains Isolated from Animals, Foods, and Humans in Spain in 1997–1998. Antimicrob. Agents Chemother. 2000, 44, 267–271. [Google Scholar] [CrossRef]
- Bremón, A.R.; Ruiz-Tovar, M.; Gorricho, B.P.; de Torres, P.D.; Rodríguez, R.L. Non-hospital consumption of antibiotics in Spain: 1987–1997. J. Antimicrob. Chemother. 2000, 45, 395–400. [Google Scholar] [CrossRef][Green Version]
- Dai, L.; Sahin, O.; Grover, M.; Zhang, Q. New and alternative strategies for the prevention, control, and treatment of antibiotic-resistant Campylobacter. Transl. Res. 2020, 223, 76–88. [Google Scholar] [CrossRef]
- Sanad, Y.M.; Closs, G.; Kumar, A.; LeJeune, J.T.; Rajashekara, G. Molecular Epidemiology and Public Health Relevance of Campylobacter Isolated from Dairy Cattle and European Starlings in Ohio, USA. Foodborne Pathog. Dis. 2013, 10, 229–236. [Google Scholar] [CrossRef]
- Wieczorek, K.; Osek, J. Antimicrobial Resistance Mechanisms among Campylobacter. Biomed Res. Int. 2013, 2013, 340605. [Google Scholar] [CrossRef]
- Iovine, N.M. Resistance Mechanisms in Campylobacter jejuni. Virulence 2013, 4, 230–240. [Google Scholar] [CrossRef]
- Hamed, S.M.; Elkhatib, W.F.; El-Mahallawy, H.A.; Helmy, M.M.; Ashour, M.S.; Khaled, M.A.; Aboshanab, K.M.A. Multiple mechanisms contributing to ciprofloxacin resistance among Gram negative bacteria causing infections to cancer patients. Sci. Rep. 2018, 8, 12268. [Google Scholar] [CrossRef]
- Garcia-Migura, L.; Hendriksen, R.S.; Fraile, L.; Aarestrup, F.M. Antimicrobial Resistance of Zoonotic and Commensal Bacteria in Europe: The Missing Link between Consumption and Resistance in Veterinary Medicine. Vet. Microbiol. 2014, 170, 1–9. [Google Scholar] [CrossRef]
- Ungemach, F.R.; Müller-Bahrdt, D.; Abraham, G. Guidelines for prudent use of antimicrobials and their implications on antibiotic usage in veterinary medicine. Int. J. Med. Microbiol. 2006, 296 (Suppl. S41), 33–38. [Google Scholar] [CrossRef]
| Sample Type | No of Samples Tested | No (%) of Samples Negative for Campylobacter | No (%) of Samples Positive for Campylobacter | No (%) of Samples Positive for C. jejuni |
|---|---|---|---|---|
| Raw milk | 12 | 12 (100%) | 0 (0%) | 0 (0%) |
| Water | 12 | 10 (83.3%) | 2 (16.7%) | 0 (0%) |
| Green grass | 23 | 21 (72.4%) | 2 (6.9%) | 0 (0%) |
| Dry forage | 11 | 11 (100%) | 0 (0%) | 0 (0%) |
| Maize cured forage | 8 | 8 (100%) | 0 (0%) | 0 (0%) |
| Grass cured forage | 5 | 5 (100%) | 0 (0%) | 0 (0%) |
| Stable floor | 105 | 101 (94.4%) | 3 (2.8%) | 1 (0.9%) |
| Slurry tanker | 47 | 44 (93.6%) | 3 (6.4%) | 0 (0%) |
| Fresh feces | 287 | 129 (53.5%) | 97 (40.2%) | 61 (25.3%) |
| Stored manure | 41 | 36 (83.7%) | 5 (11.6%) | 0 (0%) |
| Total | 551 | 377 (72.5%) | 112 (21.5%) | 62 (11.9%) |
| Antimicrobial Resistance Phenotype | No. of PFGE Type Strains |
|---|---|
| Ciprofloxacin + tetracycline | 15 |
| Ciprofloxacin | 6 |
| Total | 21 |
| Season of Collection | Type of Sample | pH | Conductivity (mS/cm) | %Dry Matter | %Ash | %Kjeldahl Nitrogen | %Ammonia Nitrogen | %Total Phosphorus |
|---|---|---|---|---|---|---|---|---|
| Autumn | 1 Fresh manure (1) | 8.30 | 4.59 | 9.5 | 2.35 | 0.27 | 0.106 | 0.050 |
| Autumn | 1 Fresh manure (2) | 7.18 | 4.45 | 11.0 | 2.37 | 0.35 | 0.097 | 0.047 |
| Spring | 1 Fresh manure (1) | 6.84 | 4.16 | 10.3 | 2.15 | 0.30 | 0.081 | 0.070 |
| Spring | 1 Fresh manure (2) | 7.76 | 3.56 | 12.3 | 3.80 | 0.36 | 0.132 | 0.066 |
| Spring | 2 Compost (1) | 6.15 | 6.86 | 9.1 | 2.80 | 0.29 | 0.159 | 0.080 |
| Spring | 2 Compost (2) | 7.57 | 5.70 | 11.8 | 4.10 | 0.35 | 0.162 | 0.086 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varsaki, A.; Ortiz, S.; Santorum, P.; López, P.; López-Alonso, V.; Martínez-Suárez, J.V. Genetic Diversity, Antimicrobial Resistance and Survival upon Manure Storage of Campylobacter jejuni Isolated from Dairy Cattle Farms in the Cantabric Coast of Spain. Zoonotic Dis. 2022, 2, 82-94. https://doi.org/10.3390/zoonoticdis2030009
Varsaki A, Ortiz S, Santorum P, López P, López-Alonso V, Martínez-Suárez JV. Genetic Diversity, Antimicrobial Resistance and Survival upon Manure Storage of Campylobacter jejuni Isolated from Dairy Cattle Farms in the Cantabric Coast of Spain. Zoonotic Diseases. 2022; 2(3):82-94. https://doi.org/10.3390/zoonoticdis2030009
Chicago/Turabian StyleVarsaki, Athanasia, Sagrario Ortiz, Patricia Santorum, Pilar López, Victoria López-Alonso, and Joaquín V. Martínez-Suárez. 2022. "Genetic Diversity, Antimicrobial Resistance and Survival upon Manure Storage of Campylobacter jejuni Isolated from Dairy Cattle Farms in the Cantabric Coast of Spain" Zoonotic Diseases 2, no. 3: 82-94. https://doi.org/10.3390/zoonoticdis2030009
APA StyleVarsaki, A., Ortiz, S., Santorum, P., López, P., López-Alonso, V., & Martínez-Suárez, J. V. (2022). Genetic Diversity, Antimicrobial Resistance and Survival upon Manure Storage of Campylobacter jejuni Isolated from Dairy Cattle Farms in the Cantabric Coast of Spain. Zoonotic Diseases, 2(3), 82-94. https://doi.org/10.3390/zoonoticdis2030009






