Dominance of Diarrheagenic E. coli Virulent Types in Integrated Crop–Livestock Farms and Their Antibiotic Resistance Patterns
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Presumptive Isolation of Diarrheagenic E. coli
2.3. Confirmation of E. coli and Identifying Their Virulent Types
2.4. Antibiotic Resistance Pattern of Confirmed Diarrheagenic E. coli Virulent Types
2.5. Statistical Analysis
3. Results
3.1. Prevalence of Diarrheagenic E. coli in Various Categories of Samples Collected at Pre- and Post-Harvest Levels
3.2. Virulent-Type-Specific Distribution of Isolated E. coli
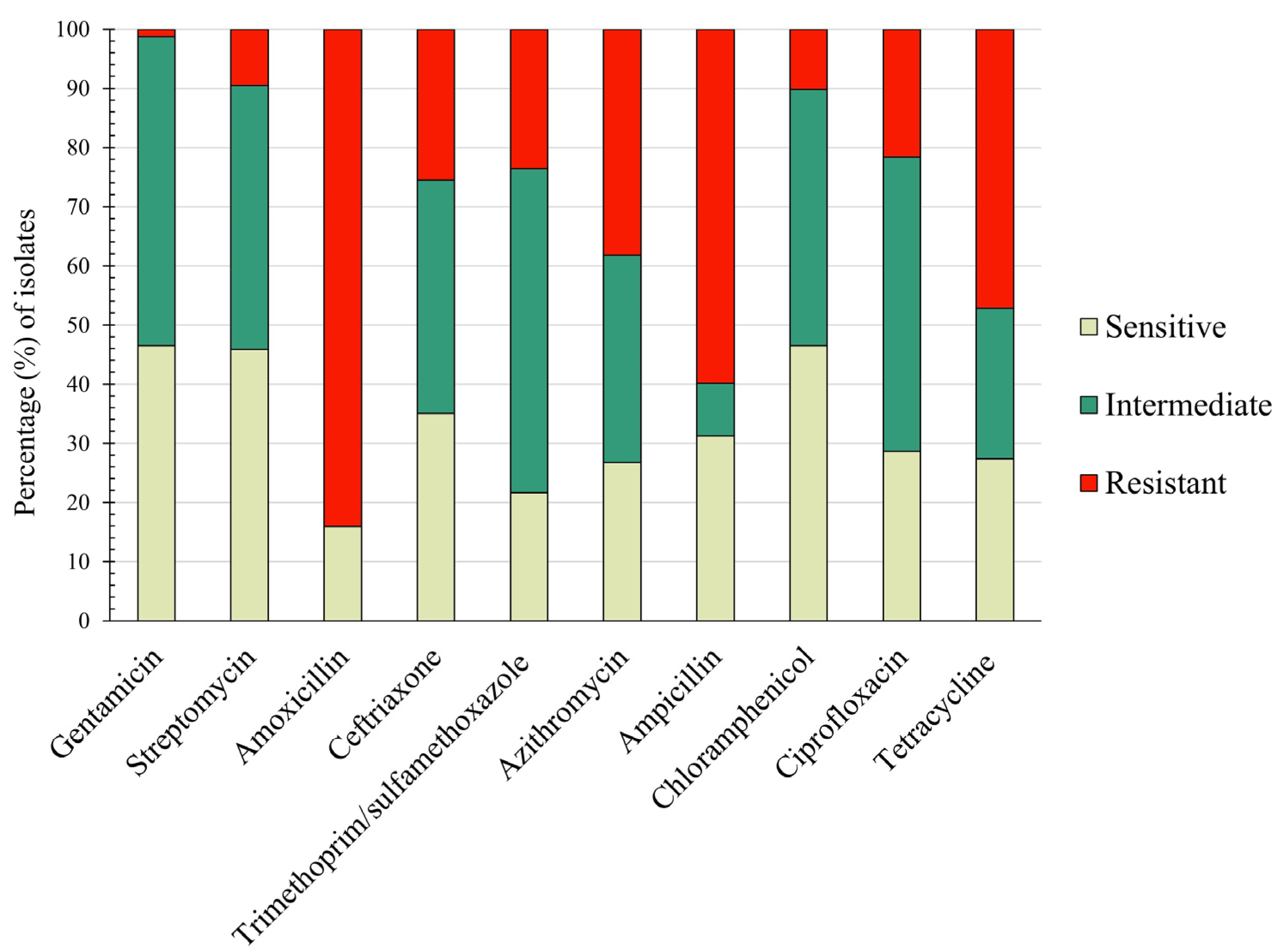
3.3. Resistance Pattern of Isolated E. coli Virulent Types against Major Antibiotics
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CDC (Centers for Disease Control and Prevention). National Outbreak Reporting System (NORS) Dashboard. 2023. Available online: https://wwwn.cdc.gov/norsdashboard/ (accessed on 1 October 2023).
- CDC (Centers for Disease Control and Prevention). Reports of Selected E. coli Outbreak Investigations. 2022. Available online: https://www.cdc.gov/ecoli/outbreaks.html (accessed on 1 October 2023).
- Tack, D.M. Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly through Food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2016–2019. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 509. [Google Scholar] [CrossRef] [PubMed]
- Ferens, W.A.; Hovde, C.J. Escherichia coli O157:H7: Animal Reservoir and Sources of Human Infection. Foodborne Pathog. Dis. 2011, 8, 465–487. [Google Scholar] [CrossRef]
- Jackson, C.R.; Davis, J.A.; Barrett, J.B. Prevalence and Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from Retail Meat and Humans in Georgia. J. Clin. Microbiol. 2013, 51, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Aditya, A.; Tabashsum, Z.; Alvarado Martinez, Z.; Wei Tung, C.; Suh, G.; Nguyen, P.; Biswas, D. Diarrheagenic Escherichia coli and Their Antibiotic Resistance Patterns in Dairy Farms and Their Microbial Ecosystems. J. Food Prot. 2023, 86, 100051. [Google Scholar] [CrossRef]
- Peng, M.; Salaheen, S.; Almario, J.A.; Tesfaye, B.; Buchanan, R.; Biswas, D. Prevalence and Antibiotic Resistance Pattern of Salmonella Serovars in Integrated Crop-Livestock Farms and Their Products Sold in Local Markets. Environ. Microbiol. 2016, 18, 1654–1665. [Google Scholar] [CrossRef]
- Painter, J.A.; Hoekstra, R.M.; Ayers, T.; Tauxe, R.V.; Braden, C.R.; Angulo, F.J.; Griffin, P.M. Attribution of Foodborne Illnesses, Hospitalizations, and Deaths to Food Commodities by Using Outbreak Data, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Berger, C.N.; Sodha, S.V.; Shaw, R.K.; Griffin, P.M.; Pink, D.; Hand, P.; Frankel, G. Fresh Fruit and Vegetables as Vehicles for the Transmission of Human Pathogens. Environ. Microbiol. 2010, 12, 2385–2397. [Google Scholar] [CrossRef] [PubMed]
- Bolton, D.J.; O’Neill, C.J.; Fanning, S. A Preliminary Study of Salmonella, Verocytotoxigenic Escherichia coli O157 and Campylobacter on Four Mixed Farms. Zoonoses Public Health 2012, 59, 217–228. [Google Scholar] [CrossRef]
- Johnston, L.M.; Jaykus, L.-A.; Moll, D.; Martinez, M.C.; Anciso, J.; Mora, B.; Moe, C.L. A Field Study of the Microbiological Quality of Fresh Produce. J. Food Prot. 2005, 68, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Kozak, G.K.; MacDonald, D.; Landry, L.; Farber, J.M. Foodborne Outbreaks in Canada Linked to Produce: 2001 through 2009. J. Food Prot. 2013, 76, 173–183. [Google Scholar] [CrossRef]
- Mukherjee, A.; Speh, D.; Jones, A.T.; Buesing, K.M.; Diez-Gonzalez, F. Longitudinal Microbiological Survey of Fresh Produce Grown by Farmers in the Upper Midwest. J. Food Prot. 2006, 69, 1928–1936. [Google Scholar] [CrossRef]
- Peng, M.; Salaheen, S.; Buchanan, R.L. Alterations of Salmonella Enterica Serovar Typhimurium Antibiotic Resistance under Environmental Pressure. Appl. Environ. Microbiol. 2018, 84, e01173-18. [Google Scholar] [CrossRef]
- Salaheen, S.; Chowdhury, N.; Hanning, I.; Biswas, D. Zoonotic Bacterial Pathogens and Mixed Crop-Livestock Farming. Poult. Sci. 2015, 94, 1398–1410. [Google Scholar] [CrossRef]
- Teramoto, H.; Salaheen, S.; Biswas, D. Contamination of Post-Harvest Poultry Products with Multidrug Resistant Staphylococcus aureus in Maryland-Washington DC Metro Area. Food Control 2016, 65, 132–135. [Google Scholar] [CrossRef]
- NFMD. National Farmers Market Directory. 2023. Available online: https://nfmd.org (accessed on 1 October 2023).
- Hoffmann, I. Climate Change and the Characterization, Breeding and Conservation of Animal Genetic Resources. Anim. Genet. 2010, 41 (Suppl. S1), 32–46. [Google Scholar] [CrossRef]
- Strawn, L.K.; Fortes, E.D.; Bihn, E.A.; Nightingale, K.K.; Gröhn, Y.T.; Worobo, R.W.; Wiedmann, M.; Bergholz, P.W. Landscape and Meteorological Factors Affecting Prevalence of Three Food-Borne Pathogens in Fruit and Vegetable Farms. Appl. Environ. Microbiol. 2013, 79, 588–600. [Google Scholar] [CrossRef]
- Barton, J.; Henderson, K. Ohio Ecological Food and Farm Association. 2008. Available online: http://certification.oeffa.org/certfiles/transition/Organic%20Transition%20Guide.pdf (accessed on 1 October 2023).
- Bullock, J.M.; Pywell, R.F.; Burke, M.J.W.; Walker, K.J. Restoration of Biodiversity Enhances Agricultural Production. Ecol. Lett. 2001, 4, 185–189. [Google Scholar] [CrossRef]
- Nielsen, M.N.; Winding, A. Microorganisms as Indicators of Soil Health. 2002. Available online: https://www2.dmu.dk/1_viden/2_publikationer/3_fagrapporter/rapporter/fr388.pdf (accessed on 1 October 2023).
- Maheux, A.F.; Picard, F.J.; Boissinot, M.; Bissonnette, L.; Paradis, S.; Bergeron, M.G. Analytical Comparison of Nine PCR Primer Sets Designed to Detect the Presence of Escherichia coli/Shigella in Water Samples. Water Res. 2009, 43, 3019–3028. [Google Scholar] [CrossRef] [PubMed]
- Botkin, D.J.; Galli, L.; Sankarapani, V.; Soler, M.; Rivas, M.; Torres, A.G. Development of a Multiplex PCR Assay for Detection of Shiga Toxin-Producing Escherichia coli, Enterohemorrhagic E. coli, and Enteropathogenic E. coli Strains. Front. Cell. Infect. Microbiol. 2012, 2, 8. [Google Scholar] [CrossRef]
- Ahmed, O.B.; Dablool, A.S. Quality Improvement of the DNA Extracted by Boiling Method in Gram Negative Bacteria. Int. J. Bioassays 2017, 6, 5347. [Google Scholar] [CrossRef]
- CLSI (Clinical and Laboratory Standards Institute). EM100 Connect—CLSI M100 ED33:2023. 2023. Available online: http://em100.edaptivedocs.net/GetDoc.aspx?doc=CLSI%20M100%20ED33:2023andscope=user (accessed on 1 October 2023).
- FDA (Food and Drug Administration). Leafy Greens STEC Action Plan. 2022. Available online: https://www.fda.gov/food/foodborne-pathogens/leafy-greens-stec-action-plan (accessed on 1 October 2023).
- Atidégla, S.C.; Huat, J.; Agbossou, E.K.; Saint-Macary, H.; Glèlè Kakai, R. Vegetable Contamination by the Fecal Bacteria of Poultry Manure: Case Study of Gardening Sites in Southern Benin. Int. J. Food Sci. 2016, 2016, e4767453. [Google Scholar] [CrossRef]
- Black, Z.; Balta, I.; Black, L.; Naughton, P.J.; Dooley, J.S.G.; Corcionivoschi, N. The Fate of Foodborne Pathogens in Manure Treated Soil. Front. Microbiol. 2021, 12, 781357. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, J.M.; Ribaudo, M.O.; Livingston, M.J.; Beckman, J.; Huang, W. Manure Use for Fertilizer and for Energy: Report to Congress. 2009. Available online: https://www.ers.usda.gov/webdocs/publications/42731/16739_ap037fm_1_.pdf?v=8487.8 (accessed on 1 October 2023).
- US EPA (United States Environmental Protection Agency). Animal Feeding Operations—Uses of Manure. National Pollutant Discharge Elimination System (NPDES). 2023. Available online: https://www.epa.gov/npdes/animal-feeding-operations-uses-manure (accessed on 1 October 2023).
- Augustin, C.; Rahman, S.Q. Composting Animal Manures: A Guide to the Process and Management of Animal Manure Compost. NDSU Extension Service. 2010. Available online: https://www.ag.ndsu.edu/manure/documents/nm1478.pdf (accessed on 1 October 2023).
- Saulo, A.A. Preventing E. coli Infection at Petting Zoos and Farm Animal Fairs. University of Hawaii at Manoa - College of Tropical Agriculture and Human Resources FST-17a. 2013. [Google Scholar]
- Lindeberg, Y.L.; Egedal, K.; Hossain, Z.Z.; Phelps, M.; Tulsiani, S.; Farhana, I.; Begum, A.; Jensen, P.K.M. Can Escherichia coli Fly? The Role of Flies as Transmitters of E. coli to Food in an Urban Slum in Bangladesh. Trop. Med. Int. Health 2018, 23, 2–9. [Google Scholar] [CrossRef]
- WHO (World Health Organization). Antibiotic Resistance. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 1 October 2023).
- Osaili, T.M.; Alaboudi, A.R.; Rahahlah, M. Prevalence and Antimicrobial Susceptibility of Escherichia coli O157:H7 on Beef Cattle Slaughtered in Amman Abattoir. Meat Sci. 2013, 93, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Gelalcha, B.D.; Ensermu, D.B.; Agga, G.E.; Vancuren, M.; Gillespie, B.E.; D’Souza, D.H.; Okafor, C.C.; Kerro Dego, O. Prevalence of Antimicrobial Resistant and Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Dairy Cattle Farms in East Tennessee. Foodborne Pathog. Dis. 2022, 19, 408–416. [Google Scholar] [CrossRef]

| Sample Category | Description | Total Sample No. |
|---|---|---|
| Livestock drinking water | Water collected from the drinking tubs of various farm animals, such as cow, pig, turkey, and chicken | 221 |
| Feces | Fresh fecal excreta of the farm animals | 266 |
| Feed | Dry feed such as hay, and salts collected from barns | 196 |
| Soil | Soil collected from various locations of farms: grazing land, produce garden, etc. | 359 |
| Bedding | Bedding material of farm animals including hay, grass, etc. | 47 |
| Grass | Grass collected from the grazing land of animals and the produce garden | 178 |
| Compost | Collected from different depths of the compost heap | 65 |
| Produce a (pre-harvest) | Aseptically collected from the garden | 885 |
| Produce (post-harvest) | Collected from organic grocery stores and a local farmers market | 756 |
| Total | 2973 |
| Genes | Primer Names | Sequences (5’-3’) | Product Sizes (bp) | References |
|---|---|---|---|---|
| uid a | uid-1 | ATGGAATTTCGCCGATTTTGC | 187 | [23] |
| uid-2 | ATTGTTTGCCTCCCTGCTGC | |||
| stx c | stx-VT1 | GAGCGAAATAATTTATATGTG | 518 | [24] |
| stx-VT2 | TGATGATGGCAATTCAGTAT | |||
| est e | est-AL1 | TTAATAGCACCCGGTACAAGCAGG | 147 | [24] |
| est-AL2 | CCTGACTCTTCAAAAGAGAAAATTAC | |||
| elt e | elt-LT1 | TCTCTATGTGCATACGGAGC | 322 | [24] |
| elt-LT2 | CCATACTGATTGCCGCAAT | |||
| ipa d | ipa-H1 | GTTCCTTGACCGCCTTTCCGATACCGTC | 619 | [24] |
| ipa-H2 | GCCGGTCAGCCACCCTCTGAGAGTAC | |||
| agg f | agg-R1 | GTATACACAAAAGAAGGAAGC | 254 | [24] |
| agg-R2 | ACAGAATCGTCAGCATCAGC | |||
| bfp b | bfp-1 | GGAAGTCAAATTCATGGGGGTAT | 300 | [24] |
| bfp-2 | GGAATCAGACGCAGACTGGTAGT | |||
| eae b,c | eae-SK1 | CCCGAATTCGGCACAAGCATAAGC | 881 | [24] |
| eae-SK2 | CCCGGATCCGTCTCGCCAGTATTCG |
| Antimicrobial Class | Antimicrobial Agent | Breakpoints (µg/mL) | ||
|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | ||
| Aminoglycosides | Gentamicin | ≥4 | ≥8 | ≥16 |
| Streptomycin | ≥16 | ≥24 | ≥32 | |
| β-Lactam | Amoxicillin | ≥8 | ≥16 | ≥32 |
| Cephems | Ceftriaxone | ≥1 | ≥2 | ≥4 |
| Folate pathway inhibitors | Trimethoprim/sulfamethoxazole | ≥2 and ≥38 | ≥3 and ≥57 | ≥4 and ≥76 |
| Macrolides | Azithromycin | ≥16 | ≥24 | ≥32 |
| Penicillin | Ampicillin | ≥8 | ≥16 | ≥32 |
| Phenicol | Chloramphenicol | ≥8 | ≥16 | ≥32 |
| Quinolones | Ciprofloxacin | ≥0.06 | ≥0.12 | ≥1 |
| Tetracyclines | Tetracycline | ≥4 | ≥8 | ≥16 |
| Sample Category | EPEC | STEC | EIEC | EAEC | ETEC |
|---|---|---|---|---|---|
| Environmental sample | 8.97% (7/78) | 16.66% (13/78) | 16.66% (13/78) | 2.56% (2/78) | 3.84% (3/78) |
| Pre-harvest | 5.12% (4/78) | 8.97% (7/78) | 29.48% (23/78) | 5.12% (4/78) | 2.56% (2/78) |
| Post-harvest | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Sources | EPEC | STEC | EIEC | EAEC | ETEC | Distribution (%) |
|---|---|---|---|---|---|---|
| Water | 3 | 3 | 8 | 1 | 0 | 19.23 |
| Feed | 2 | 5 | 2 | 0 | 0 | 11.53 |
| Feces | 0 | 1 | 1 | 0 | 3 | 6.41 |
| Bedding | 0 | 1 | 0 | 0 | 0 | 1.28 |
| Soil | 1 | 0 | 2 | 1 | 0 | 5.12 |
| Grass | 1 | 3 | 0 | 0 | 0 | 5.12 |
| Pre-harvest produce | 4 | 7 | 23 | 4 | 2 | 51.28 |
| Post-harvest produce | 0 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aditya, A.; Julianingsih, D.; Tabashsum, Z.; Alvarado-Martinez, Z.; Tung, C.-W.; Wall, M.; Biswas, D. Dominance of Diarrheagenic E. coli Virulent Types in Integrated Crop–Livestock Farms and Their Antibiotic Resistance Patterns. Zoonotic Dis. 2024, 4, 11-21. https://doi.org/10.3390/zoonoticdis4010003
Aditya A, Julianingsih D, Tabashsum Z, Alvarado-Martinez Z, Tung C-W, Wall M, Biswas D. Dominance of Diarrheagenic E. coli Virulent Types in Integrated Crop–Livestock Farms and Their Antibiotic Resistance Patterns. Zoonotic Diseases. 2024; 4(1):11-21. https://doi.org/10.3390/zoonoticdis4010003
Chicago/Turabian StyleAditya, Arpita, Dita Julianingsih, Zajeba Tabashsum, Zabdiel Alvarado-Martinez, Chuan-Wei Tung, Matthew Wall, and Debabrata Biswas. 2024. "Dominance of Diarrheagenic E. coli Virulent Types in Integrated Crop–Livestock Farms and Their Antibiotic Resistance Patterns" Zoonotic Diseases 4, no. 1: 11-21. https://doi.org/10.3390/zoonoticdis4010003
APA StyleAditya, A., Julianingsih, D., Tabashsum, Z., Alvarado-Martinez, Z., Tung, C.-W., Wall, M., & Biswas, D. (2024). Dominance of Diarrheagenic E. coli Virulent Types in Integrated Crop–Livestock Farms and Their Antibiotic Resistance Patterns. Zoonotic Diseases, 4(1), 11-21. https://doi.org/10.3390/zoonoticdis4010003






