The G Protein-Coupled Estrogen Receptor GPER in the Development and Progression of Cancer
Abstract
1. Introduction
2. GPER Characteristics
2.1. GPER Structure and Binding Modes
2.2. GPER Ligands
2.2.1. Canonical GPER Ligands
2.2.2. Non-Canonical GPER Ligands
2.3. GPER Signaling Pathways in Cancer Cells
2.4. GPER Signaling Pathways in Cancer Chemorresistance
3. Role of GPER in Human Cancer
3.1. In Silico Studies of Different Cancer Types
3.2. Breast Cancer
3.3. Cervical Cancer
3.4. Colorectal Cancer
3.5. Endometrial Cancer
3.6. Gastric Cancer
3.7. Hepatocellular Carcinoma
3.8. Leukemia and Lymphoma
3.9. Lung Cancer
3.10. Ovarian Cancer
3.11. Thyroid Cancer
3.12. Other Cancers
4. Role of GPER in the Tumor Microenvironment
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, P.; Li, B.; Ou-Yang, L. Role of Estrogen Receptors in Health and Disease. Front. Endocrinol. 2022, 13, 839005. [Google Scholar] [CrossRef] [PubMed]
- Carmeci, C.; Thompson, D.A.; Ring, H.Z.; Francke, U.; Weigel, R.J. Identification of a Gene (GPR30) with Homology to the G-Protein-Coupled Receptor Superfamily Associated with Estrogen Receptor Expression in Breast Cancer. Genomics 1997, 617, 607–617. [Google Scholar] [CrossRef]
- Prossnitz, E.R.; Barton, M. The G-Protein-Coupled Estrogen Receptor GPER in Health and Disease. Nat. Rev. Endocrinol. 2011, 7, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Prossnitz, E.R.; Barton, M. The G Protein-Coupled Oestrogen Receptor GPER in Health and Disease: An Update. Nat. Rev. Endocrinol. 2023, 19, 407–424. [Google Scholar] [CrossRef] [PubMed]
- Ariazi, E.A.; Brailoiu, E.; Yerrum, S.; Shupp, H.A.; Slifker, M.J.; Cunliffe, H.E.; Black, M.A.; Donato, A.L.; Arterburn, J.B.; Oprea, T.I.; et al. The G Protein-Coupled Receptor GPR30 Inhibits Proliferation of Estrogen Receptor-Positive Breast Cancer Cells. Cancer Res. 2010, 70, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Chen, Z.J.; Zhang, K.S.; Yang, X.L.; Wu, Y.M.; Chen, X.H.; Huang, H.B.; Liu, H.L.; Cai, S.H.; Du, J.; et al. The Activation of G Protein-Coupled Receptor 30 (GPR30) Inhibits Proliferation of Estrogen Receptornegative Breast Cancer Cells In Vitro and In Vivo. Cell Death Dis. 2014, 5, e1428. [Google Scholar] [CrossRef] [PubMed]
- Weißenborn, C.; Ignatov, T.; Ochel, H.J.; Costa, S.D.; Zenclussen, A.C.; Ignatova, Z.; Ignatov, A. GPER Functions as a Tumor Suppressor in Triple-Negative Breast Cancer Cells. J. Cancer Res. Clin. Oncol. 2014, 140, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Barton, M.; Filardo, E.J.; Lolait, S.J.; Thomas, P.; Maggiolini, M.; Prossnitz, E.R. Twenty Years of the G Protein-Coupled Estrogen Receptor GPER: Historical and Personal Perspectives. J. Steroid Biochem. Mol. Biol. 2018, 176, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Jung, J. Role of G Protein-Coupled Estrogen Receptor in Tamoxifen-Resistant Breast Cancer MCF-7 Cells. Toxicol. Res. 2019, 35, 209–214. [Google Scholar] [CrossRef]
- Pepermans, R.A.; Sharma, G.; Prossnitz, E.R. G Protein-Coupled Estrogen Receptor in Cancer and Stromal Cells: Functions and Novel Therapeutic Perspectives. Cells 2021, 10, 672. [Google Scholar] [CrossRef]
- Hall, K.A.; Filardo, E.J. The G Protein-Coupled Estrogen Receptor (GPER): A Critical Therapeutic Target for Cancer. Cells 2023, 12, 2460. [Google Scholar] [CrossRef] [PubMed]
- Arterburn, J.B.; Prossnitz, E.R. G Protein-Coupled Estrogen Receptor GPER: Molecular Pharmacology and Therapeutic Applications. Annu. Rev. Pharmacol. Toxicol. 2023, 63, 295–320. [Google Scholar] [CrossRef] [PubMed]
- Jala, V.R.; Radde, B.N.; Haribabu, B.; Klinge, C.M. Enhanced Expression of G-Protein Coupled Estrogen Receptor (GPER/GPR30) in Lung Cancer. BMC Cancer 2012, 12, 624. [Google Scholar] [CrossRef] [PubMed]
- Aiello, F.; Carullo, G.; Giordano, F.; Spina, E.; Nigro, A.; Garofalo, A.; Tassini, S.; Costantino, G.; Vincetti, P.; Bruno, A.; et al. Identification of Breast Cancer Inhibitors Specific for G Protein-Coupled Estrogen Receptor (GPER)-Expressing Cells. ChemMedChem 2017, 12, 1279–1285. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.; Aiello, F.; Costantino, G.; Radi, M. Homology Modeling, Validation and Dynamics of the G Protein-Coupled Estrogen Receptor 1 (GPER-1). Mol. Inform. 2016, 35, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Arnatt, C.K.; Zhang, Y. G Protein-Coupled Estrogen Receptor (GPER) Agonist Dual Binding Mode Analyses toward Understanding of Its Activation Mechanism: A Comparative Homology Modeling Approach. Mol. Inform. 2013, 32, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.U.; Ahemad, N.; Chuah, L.H.; Naidu, R.; Htar, T.T. Sequential Ligand- and Structure-Based Virtual Screening Approach for the Identification of Potential G Protein-Coupled Estrogen Receptor-1 (GPER-1) Modulators. RSC Adv. 2019, 9, 2525–2538. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Luna, D.; Martínez-Archundia, M.; Maroun, R.C.; Ceballos-Reyes, G.; Fragoso-Vázquez, M.J.; González-Juárez, D.E.; Correa-Basurto, J. Deciphering the GPER/GPR30-Agonist and Antagonists Interactions Using Molecular Modeling Studies, Molecular Dynamics, and Docking Simulations. J. Biomol. Struct. Dyn. 2015, 33, 2161–2172. [Google Scholar] [CrossRef]
- Bello, M.; Méndez-Luna, D.; Sarmiento, V.; Correa Basurto, J.; Najera, N.; Villarreal, F.; Ceballos, G. Structural and Energetic Basis for Novel Epicatechin Derivatives Acting as GPER Agonists through the MMGBSA Method. J. Steroid Biochem. Mol. Biol. 2019, 189, 176–186. [Google Scholar] [CrossRef]
- Sarmiento, V.; Ramirez-Sanchez, I.; Moreno-Ulloa, A.; Romero-Perez, D.; Chávez, D.; Ortiz, M.; Najera, N.; Correa-Basurto, J.; Villarreal, F.; Ceballos, G. Synthesis of Novel (−)-Epicatechin Derivatives as Potential Endothelial GPER Agonists: Evaluation of Biological Effects. Bioorganic Med. Chem. Lett. 2018, 28, 658–663. [Google Scholar] [CrossRef]
- Bologa, C.G.; Revankar, C.M.; Young, S.M.; Edwards, B.S.; Arterburn, J.B.; Kiselyov, A.S.; Parker, M.A.; Tkachenko, S.E.; Savchuck, N.P.; Sklar, L.A.; et al. Virtual and Biomolecular Screening Converge on a Selective Agonist for GPR30. Nat. Chem. Biol. 2006, 2, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Dennis, M.K.; Burai, R.; Ramesh, C.; Petrie, W.K.; Alcon, S.N.; Nayak, T.K.; Bologa, C.G.; Leitao, A.; Brailoiu, E.; Deliu, E.; et al. In Vivo Effects of a GPR30 Antagonist. Nat. Chem. Biol. 2009, 5, 421–427. [Google Scholar] [CrossRef]
- Dennis, M.K.; Field, A.S.; Burai, R.; Ramesh, C.; Petrie, W.K.; Bologa, C.G.; Oprea, T.I.; Yamaguchi, Y.; Hayashi, S.; Sklar, L.A.; et al. Identification of a GPER/GPR30 Antagonist with Improved Estrogen Receptor Counterselectivity. J. Steroid Biochem. Mol. Biol. 2011, 127, 358–366. [Google Scholar] [CrossRef]
- O’Dea, A.; Sondergard, C.; Sweeney, P.; Arnatt, C.K. A Series of Indole-Thiazole Derivatives Act as GPER Agonists and Inhibit Breast Cancer Cell Growth. ACS Med. Chem. Lett. 2018, 9, 901–906. [Google Scholar] [CrossRef]
- Burai, R.; Ramesh, C.; Nayak, T.K.; Dennis, M.K.; Bryant, B.K.; Prossnitz, E.R.; Arterburn, J.B. Synthesis and Characterization of Tricarbonyl-Re/Tc(I) Chelate Probes Targeting the G Protein-Coupled Estrogen Receptor GPER/GPR30. PLoS ONE 2012, 7, e46861. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Zhang, X.; Xie, Y.; Tu, Y.; Wang, D.; Liu, Z.; Wang, Z.Y. Involvement of Estrogen Receptor Variant ER-A36, Not GPR30, in Nongenomic Estrogen Signaling. Mol. Endocrinol. 2010, 24, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; He, C.; Huang, C.; Hua, G.; Wang, Z.; Remmenga, S.W.; Rodabough, K.J.; Karpf, A.R.; Dong, J.; Davis, J.S.; et al. G-1 Inhibits Breast Cancer Cell Growth via Targeting Colchicine-Binding Site of Tubulin to Interfere with Microtubule Assembly. Mol. Cancer Ther. 2017, 16, 1080–1091. [Google Scholar] [CrossRef]
- Wang, C.; Lv, X.; He, C.; Hua, G.; Tsai, M.-Y.; Davis, J.S. The G-Protein-Coupled Estrogen Receptor Agonist G-1 Suppresses Proliferation of Ovarian Cancer Cells by Blocking Tubulin Polymerization. Cell Death Dis. 2013, 4, e869. [Google Scholar] [CrossRef]
- Hirtz, A.; Lebourdais, N.; Rech, F.; Bailly, Y.; Vaginay, A.; Smaïl-Tabbone, M.; Dubois-Pot-schneider, H.; Dumond, H. Gper Agonist G-1 Disrupts Tubulin Dynamics and Potentiates Temozolomide to Impair Glioblastoma Cell Proliferation. Cells 2021, 10, 3438. [Google Scholar] [CrossRef]
- Torres-López, L.; Olivas-Aguirre, M.; Villatoro-Gómez, K.; Dobrovinskaya, O. The G-Protein–Coupled Estrogen Receptor Agonist G-1 Inhibits Proliferation and Causes Apoptosis in Leukemia Cell Lines of T Lineage. Front. Cell Dev. Biol. 2022, 10, 811479. [Google Scholar] [CrossRef]
- Liu, Y.; Du, F.-Y.; Chen, W.; Fu, P.-F.; Yao, M.-Y.; Zheng, S.-S. G15 Sensitizes Epithelial Breast Cancer Cells to Doxorubicin by Preventing Epithelial-Mesenchymal Transition through Inhibition of GPR30. Am. J. Transl. Res. 2015, 7, 967–975. [Google Scholar] [PubMed]
- Wang, X.; Xu, Z.; Sun, J.; Lv, H.; Wang, Y.; Ni, Y.; Chen, S.; Hu, C.; Wang, L.; Chen, W.; et al. Cisplatin Resistance in Gastric Cancer Cells Is Involved with GPR30-Mediated Epithelial-Mesenchymal Transition. J. Cell. Mol. Med. 2020, 24, 3625–3633. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.Y.; Weng, J.R.; Hu, J.L.; Wang, D.; Sargeant, A.M.; Chiu, C.F. G15, a GPR30 Antagonist, Induces Apoptosis and Autophagy in Human Oral Squamous Carcinoma Cells. Chem. Biol. Interact. 2013, 206, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Blesson, C.S.; Sahlin, L. Expression Pattern and Signalling Pathways in Neutrophil like HL-60 Cells after Treatment with Estrogen Receptor Selective Ligands. Mol. Cell. Endocrinol. 2012, 361, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Heublein, S.; Jeschke, U.; Kuhn, C.; Hester, A.; Czogalla, B.; Mahner, S.; Rottmann, M.; Mayr, D.; Schmoeckel, E.; et al. The G-Protein-Coupled Estrogen Receptor (GPER) Regulates Trimethylation of Histone H3 at Lysine 4 and Represses Migration and Proliferation of Ovarian Cancer Cells In Vitro. Cells 2021, 10, 619. [Google Scholar] [CrossRef] [PubMed]
- Girgert, R.; Emons, G.; Gründker, C. Inactivation of GPR30 Reduces Growth of Triple-Negative Breast Cancer Cells: Possible Application in Targeted Therapy. Breast Cancer Res. Treat. 2012, 134, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Vivacqua, A.; Bonofiglio, D.; Albanito, L.; Madeo, A.; Rago, V.; Carpino, A.; Musti, A.M.; Picard, D.; Andò, S.; Maggiolini, M. 17β-Estradiol, Genistein, and 4-Hydroxytamoxifen Induce the Proliferation of Thyroid Cancer Cells through the G Protein-Coupled Receptor GPR30. Mol. Pharmacol. 2006, 70, 1414–1423. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Y.; Lan, L.; Liu, R.; Wu, Y.; Qu, Q.; Wen, K. Tamoxifen Has a Proliferative Effect in Endometrial Carcinoma Mediated via the GPER/EGFR/ERK/Cyclin D1 Pathway: A Retrospective Study and an in Vitro Study. Mol. Cell. Endocrinol. 2016, 437, 51–61. [Google Scholar] [CrossRef]
- Tsai, C.L.; Wu, H.M.; Lin, C.Y.; Lin, Y.J.; Chao, A.; Wang, T.H.; Hsueh, S.; Lai, C.H.; Wang, H.S. Estradiol and Tamoxifen Induce Cell Migration through GPR30 and Activation of Focal Adhesion Kinase (FAK) in Endometrial Cancers with Low or without Nuclear Estrogen Receptor α (ERα). PLoS ONE 2013, 8, e72999. [Google Scholar] [CrossRef]
- Avena, P.; Casaburi, I.; Zavaglia, L.; Nocito, M.C.; La Padula, D.; Rago, V.; Dong, J.; Thomas, P.; Mineo, C.; Sirianni, R.; et al. 27-Hydroxycholesterol Binds GPER and Induces Progression of Estrogen Receptor-Negative Breast Cancer. Cancers 2022, 14, 1521. [Google Scholar] [CrossRef]
- Castillo Sanchez, R.; Gomez, R.; Perez Salazar, E. Bisphenol A Induces Migration through a GPER-, FAK-, Src-, and ERK2-Dependent Pathway in MDA-MB-231 Breast Cancer Cells. Chem. Res. Toxicol. 2016, 29, 285–295. [Google Scholar] [CrossRef]
- Zhu, P.; Liao, L.Y.; Zhao, T.T.; Mo, X.M.; Chen, G.G.; Liu, Z.M. GPER/ERK&AKT/NF-ΚB Pathway Is Involved in Cadmium-Induced Proliferation, Invasion and Migration of GPER-Positive Thyroid Cancer Cells. Mol. Cell. Endocrinol. 2017, 442, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Filardo, E.J.; Shaikh, Z.A. The Membrane Estrogen Receptor GPR30 Mediates Cadmium-Induced Proliferation of Breast Cancer Cells. Toxicol. Appl. Pharmacol. 2010, 245, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Catusse, J.; Wollner, S.; Leick, M.; Schröttner, P.; Schraufstätter, I.; Burger, M. Attenuation of CXCR4 Responses by CCL18 in Acute Lymphocytic Leukemia B Cells. J. Cell. Physiol. 2010, 225, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Radde, B.N.; Litchfield, L.M.; Ivanova, M.M.; Prough, R.A.; Clark, B.J.; Doll, M.A.; Hein, D.W.; Klinge, C.M. Dehydroepiandrosterone Activation of G-Protein-Coupled Estrogen Receptor Rapidly Stimulates MicroRNA-21 Transcription in Human Hepatocellular Carcinoma Cells. J. Biol. Chem. 2015, 290, 15799–15811. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, M.F.; Yang, M.L.; Wu, T.Y.; Xu, C.J.; Wang, J.M.; Li, C.J.; Li, X. G Protein-Coupled Receptor 30 Mediates the Anticancer Effects Induced by Eicosapentaenoic Acid in Ovarian Cancer Cells. Cancer Res. Treat. 2020, 52, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Jouffre, B.; Acramel, A.; Belnou, M.; Santolla, M.F.; Talia, M.; Lappano, R.; Nemati, F.; Decaudin, D.; Khemtemourian, L.; Liu, W.Q.; et al. Identification of a Human Estrogen Receptor α Tetrapeptidic Fragment with Dual Antiproliferative and Anti-Nociceptive Action. Sci. Rep. 2023, 13, 1326. [Google Scholar] [CrossRef] [PubMed]
- Lappano, R.; Mallet, C.; Rizzuti, B.; Grande, F.; Galli, G.; Byrne, C.; Broutin, I.; Boudieu, L.; Eschalier, A.; Jacquot, Y.; et al. The Peptide ERα17p Is a GPER Inverse Agonist That Exerts Antiproliferative Effects in Breast Cancer Cells. Cells 2019, 8, 590. [Google Scholar] [CrossRef] [PubMed]
- Lappano, R.; Rosano, C.; De Marco, P.; De Francesco, E.M.; Pezzi, V.; Maggiolini, M. Estriol Acts as a GPR30 Antagonist in Estrogen Receptor-Negative Breast Cancer Cells. Mol. Cell. Endocrinol. 2010, 320, 162–170. [Google Scholar] [CrossRef]
- Giessrigl, B.; Schmidt, W.M.; Kalipciyan, M.; Jeitler, M.; Bilban, M.; Gollinger, M.; Krieger, S.; Jäger, W.; Mader, R.M.; Krupitza, G. Fulvestrant Induces Resistance by Modulating GPER and CDK6 Expression: Implication of Methyltransferases, Deacetylases and the HSWI/SNF Chromatin Remodelling Complex. Br. J. Cancer 2013, 109, 2751–2762. [Google Scholar] [CrossRef]
- Lappano, R.; Rosano, C.; Santolla, M.F.; Pupo, M.; De Francesco, E.M.; De Marco, P.; Ponassi, M.; Spallarossa, A.; Ranise, A.; Maggiolini, M. Two Novel GPER Agonists Induce Gene Expression Changes and Growth Effects in Cancer Cells. Curr. Cancer Drug Targets 2012, 12, 531–542. [Google Scholar] [CrossRef]
- Chimento, A.; Casaburi, I.; Rosano, C.; Avena, P.; De Luca, A.; Campana, C.; Martire, E.; Santolla, M.F.; Maggiolini, M.; Pezzi, V.; et al. Oleuropein and Hydroxytyrosol Activate GPER/GPR30-Dependent Pathways Leading to Apoptosis of ER-Negative SKBR3 Breast Cancer Cells. Mol. Nutr. Food Res. 2014, 58, 478–489. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Tan, W.; Zheng, J.; Zhang, B.; Li, H.; Li, X. MEHP Promotes the Proliferation of Cervical Cancer via GPER Mediated Activation of Akt. Eur. J. Pharmacol. 2018, 824, 11–16. [Google Scholar] [CrossRef]
- Lappano, R.; Santolla, M.F.; Pupo, M.; Sinicropi, M.S.; Caruso, A.; Rosano, C.; Maggiolini, M. MIBE Acts as Antagonist Ligand of Both Estrogen Receptor α and GPER in Breast Cancer Cells. Breast Cancer Res. 2012, 14, R12. [Google Scholar] [CrossRef] [PubMed]
- Santolla, M.F.; De Francesco, E.M.; Lappano, R.; Rosano, C.; Abonante, S.; Maggiolini, M. Niacin Activates the G Protein Estrogen Receptor (GPER)-Mediated Signalling. Cell. Signal. 2014, 26, 1466–1475. [Google Scholar] [CrossRef] [PubMed]
- Maggiolini, M.; Santolla, M.F.; Avino, S.; Aiello, F.; Rosano, C.; Garofalo, A.; Grande, F. Identification of Two Benzopyrroloxazines Acting as Selective GPER Antagonists in Breast Cancer Cells and Cancer-Associated Fibroblasts. Future Med. Chem. 2015, 7, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Zekas, E.; Prossnitz, E.R. Estrogen-Mediated Inactivation of FOXO3a by the G Protein-Coupled Estrogen Receptor GPER. BMC Cancer 2015, 15, 702. [Google Scholar] [CrossRef]
- Ignatov, A.; Ignatov, T.; Roessner, A.; Costa, S.D.; Kalinski, T. Role of GPR30 in the Mechanisms of Tamoxifen Resistance in Breast Cancer MCF-7 Cells. Breast Cancer Res. Treat. 2010, 123, 87–96. [Google Scholar] [CrossRef]
- Yu, T.; Cheng, H.; Ding, Z.; Wang, Z.; Zhou, L.; Zhao, P.; Tan, S.; Xu, X.; Huang, X.; Liu, M.; et al. GPER Mediates Decreased Chemosensitivity via Regulation of ABCG2 Expression and Localization in Tamoxifen-Resistant Breast Cancer Cells. Mol. Cell. Endocrinol. 2020, 506, 110762. [Google Scholar] [CrossRef]
- Ignatov, T.; Eggemann, H.; Semczuk, A.; Smith, B.; Bischoff, J.; Roessner, A.; Costa, S.D.; Kalinski, T.; Ignatov, A. Role of GPR30 in Endometrial Pathology after Tamoxifen for Breast Cancer. Am. J. Obstet. Gynecol. 2010, 203, 595.e9–595.e16. [Google Scholar] [CrossRef]
- Yuan, J.; Liu, M.; Yang, L.; Gang, T.; Zhu, Q.; Chen, M.; Chen, H.; Luo, H.; Fu, Q.; Li, Z.; et al. Acquisition of epithelial-mesenchymal transition phenotype in the tamoxifen-resistant breast cancer cell: A new role for G protein-coupled estrogen receptor in mediating tamoxifen resistance through cancer-associated fibroblast-derived fibronectin and β1-integrin signaling pathway in tumor cells. Breast Cancer Res. 2015, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Schutyser, E.; Richmond, A.; Van Damme, J. Involvement of CC Chemokine Ligand 18 (CCL18) in Normal and Pathological Processes. J. Leukoc. Biol. 2005, 78, 14–26. [Google Scholar] [CrossRef]
- Schmidt-Wolf, R.; Zissel, G. Interaction Between CCL18 and GPR30 Differs from the Interaction Between Estradiol and GPR30. Anticancer Res. 2020, 40, 3097–3108. [Google Scholar] [CrossRef] [PubMed]
- Tuli, H.S.; Tuorkey, M.J.; Thakral, F.; Sak, K.; Kumar, M.; Sharma, A.K.; Sharma, U.; Jain, A.; Aggarwal, V.; Bishayee, A. Molecular Mechanisms of Action of Genistein in Cancer: Recent Advances. Front. Pharmacol. 2019, 10, 1336. [Google Scholar] [CrossRef] [PubMed]
- Hafezi, S.A.; Abdel-Rahman, W.M. The Endocrine Disruptor Bisphenol A (BPA) Exerts a Wide Range of Effects in Carcinogenesis and Response to Therapy. Curr. Mol. Pharmacol. 2019, 12, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Bimonte, V.M.; Besharat, Z.M.; Antonioni, A.; Cella, V.; Lenzi, A.; Ferretti, E.; Migliaccio, S. The Endocrine Disruptor Cadmium: A New Player in the Pathophysiology of Metabolic Diseases. J. Endocrinol. Investig. 2021, 44, 1363–1377. [Google Scholar] [CrossRef]
- Bimonte, V.M.; Catanzaro, G.; Po, A.; Trocchianesi, S.; Besharat, Z.M.; Spinello, Z.; Curreli, M.; Fabi, A.; Bei, R.; Milella, M.; et al. The Endocrine Disruptor Cadmium Modulates the Androgen–Estrogen Receptors Ratio and Induces Inflammatory Cytokines in Luminal (A) Cell Models of Breast Cancer. Endocrine 2023, 83, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Kampa, M.; Lappano, R.; Grande, F.; Rizzuti, B.; Maggiolini, M.; Castanas, E.; Jacquot, Y. Promising Perspectives of the Antiproliferative GPER Inverse Agonist ERα17p in Breast Cancer. Cells 2023, 12, 653. [Google Scholar] [CrossRef] [PubMed]
- Filardo, E.J.; Quinn, J.A.; Bland, K.I.; Frackelton, A.R., Jr. Estrogen-Induced Activation of Erk-1 and Erk-2 Requires the G Protein-Coupled Receptor Homolog, GPR30, and Occurs via Trans-Activation of the Epidermal Growth Factor Receptor through Release of HB-EGF. Mol. Endocrinol. 2000, 14, 1649–1660. [Google Scholar] [CrossRef]
- Filardo, E.J.; Thomas, P. GPR30: A Seven-Transmembrane-Spanning Estrogen Receptor That Triggers EGF Release. Trends Endocrinol. Metab. 2005, 16, 362–367. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-J.; Pan, W.-W.; Liu, S.B.; Shen, Z.-F.; Xu, Y.; Hu, L.L. ERK/MAPK Signalling Pathway and Tumorigenesis (Review). Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT Signaling Pathway: From Bench to Clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.K. MAP Kinase Pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254. [Google Scholar] [CrossRef] [PubMed]
- Filardo, E.J.; Quinn, J.A.; Frackelton, A.R.; Bland, K.I.; Endocrin, M. Estrogen Action Via the G Protein-Coupled Receptor, GPR30: Stimulation of Adenylyl Cyclase and CAMP-Mediated Attenuation of the Epidermal Growth Factor Receptor-to-MAPK Signaling Axis. Mol. Endocrinol. 2002, 16, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.F.; Wu, T.T.; Yang, J.Y.; Dong, C.R.; Wang, N.; Liu, X.H.; Liu, Z.M. 17β-Estradiol Promotes the Invasion and Migration of Nuclear Estrogen Receptor-Negative Breast Cancer Cells through Cross-Talk between GPER1 and CXCR1. J. Steroid Biochem. Mol. Biol. 2013, 138, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Santolla, M.F.; Lappano, R.; De Marco, P.; Pupo, M.; Vivacqua, A.; Sisci, D.; Abonante, S.; Iacopetta, D.; Cappello, A.R.; Dolce, V.; et al. G Protein-Coupled Estrogen Receptor Mediates the Up-Regulation of Fatty Acid Synthase Induced by 17β–Estradiol in Cancer Cells and Cancer-Associated Fibroblasts. J. Biol. Chem. 2012, 287, 43234–43245. [Google Scholar] [CrossRef] [PubMed]
- Vivacqua, A.; Sebastiani, A.; Miglietta, A.M.; Rigiracciolo, D.C.; Cirillo, F.; Galli, G.R.; Talia, M.; Santolla, M.F.; Lappano, R.; Giordano, F.; et al. MiR-338-3p Is Regulated by Estrogens through GPER in Breast Cancer Cells and Cancer-Associated Fibroblasts (CAFs). Cells 2018, 7, 203. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Jiang, X.; Zhao, Y.; Wen, H.; Liu, G. Role of GPER on Proliferation, Migration and Invasion in Ligand-Independent Manner in Human Ovarian Cancer Cell Line SKOV3. Cell Biochem. Funct. 2015, 33, 552–559. [Google Scholar] [CrossRef]
- Milde-Langosch, K. The Fos Family of Transcription Factors and Their Role in Tumourigenesis. Eur. J. Cancer 2005, 41, 2449–2461. [Google Scholar] [CrossRef]
- Muhammad, N.; Bhattacharya, S.; Steele, R.; Phillips, N.; Ray, R.B. Involvement of C-Fos in the Promotion of Cancer Stem-like Cell Properties in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2017, 23, 3120–3128. [Google Scholar] [CrossRef] [PubMed]
- Farhan, M.; Wang, H.; Gaur, U.; Little, P.J.; Xu, J.; Zheng, W. FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int. J. Biol. Sci. 2017, 13, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ao, X.; Ding, W.; Ponnusamy, M.; Wu, W.; Hao, X.; Yu, W.; Wang, Y.; Li, P.; Wang, J. Critical Role of FOXO3a in Carcinogenesis. Mol. Cancer 2018, 17, 104. [Google Scholar] [CrossRef] [PubMed]
- Wallacides, A.; Chesnel, A.; Ajj, H.; Chillet, M.; Flament, S.; Dumond, H. Estrogens Promote Proliferation of the Seminoma-like TCam-2 Cell Line through a GPER-Dependent ERα36 Induction. Mol. Cell. Endocrinol. 2012, 350, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Notas, G.; Panagiotopoulos, A.; Vamvoukaki, R.; Kalyvianaki, K.; Kiagiadaki, F.; Deli, A.; Kampa, M.; Castanas, E. Erα36–Gper1 Collaboration Inhibits Tlr4/Nfκb-Induced pro-Inflammatory Activity in Breast Cancer Cells. Int. J. Mol. Sci. 2021, 22, 7603. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The Basics of Epithelial-Mesenchymal Transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhu, X.Y.; Jiang, R.; Xu, M.; Wang, N.; Chen, G.G.; Liu, Z.M. Role of GPER1, EGFR and CXCR1 in Differentiating between Malignant Follicular Thyroid Carcinoma and Benign Follicular Thyroid Adenoma. Int. J. Clin. Exp. Pathol. 2015, 8, 11236–11247. [Google Scholar] [PubMed]
- Xu, E.; Xia, X.; Jiang, C.; Li, Z.; Yang, Z.; Zheng, C.; Wang, X.; Du, S.; Miao, J.; Wang, F.; et al. GPER1 Silencing Suppresses the Proliferation, Migration, and Invasion of Gastric Cancer Cells by Inhibiting PI3K/AKT–Mediated EMT. Front. Cell Dev. Biol. 2020, 8, 591239. [Google Scholar] [CrossRef] [PubMed]
- Tirado-Garibay, A.C.; Falcón-Ruiz, E.A.; Ochoa-Zarzosa, A.; López-Meza, J.E. GPER: An Estrogen Receptor Key in Metastasis and Tumoral Microenvironments. Int. J. Mol. Sci. 2023, 24, 14993. [Google Scholar] [CrossRef]
- Rice, A.; Cortes, E.; Lachowski, D.; Oertle, P.; Matellan, C.; Thorpe, S.D.; Ghose, R.; Wang, H.; Lee, D.A.; Plodinec, M.; et al. GPER Activation Inhibits Cancer Cell Mechanotransduction and Basement Membrane Invasion via RhoA. Cancers 2020, 12, 289. [Google Scholar] [CrossRef]
- Ruckriegl, S.; Loris, J.; Wert, K.; Bauerschmitz, G.; Gallwas, J.; Grundker, C. Knockdown of G Protein-Coupled Estrogen Receptor 1 (GPER1) Enhances Tumor-Supportive Properties in Cervical Carcinoma Cells. Cancer Genom. Proteom. 2023, 20, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Li, C.; Zhou, L.; Huang, J. G Protein-coupled Oestrogen Receptor Promotes Cell Growth of Non-small Cell Lung Cancer Cells via YAP1/QKI/CircNOTCH1/M6A Methylated NOTCH1 Signalling. J. Cell. Mol. Med. 2021, 25, 284–296. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Wang, Z.; Li, H.; Shou, J.; Jing, Z.; Xie, J.; Sui, X.; Pan, H.; Han, W. The Role of STAT3 in Autophagy. Autophagy 2015, 11, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Santolla, M.F.; Avino, S.; Pellegrino, M.; De Francesco, E.M.; De Marco, P.; Lappano, R.; Vivacqua, A.; Cirillo, F.; Rigiracciolo, D.C.; Scarpelli, A.; et al. SIRT1 Is Involved in Oncogenic Signaling Mediated by GPER in Breast Cancer. Cell Death Dis. 2015, 6, e1834. [Google Scholar] [CrossRef]
- Lee, I.H.; Cao, L.; Mostoslavsky, R.; Lombard, D.B.; Liu, J.; Bruns, N.E.; Tsokos, M.; Alt, F.W.; Finkel, T. A Role for the NAD-Dependent Deacetylase Sirt1 in the Regulation of Autophagy. Proc. Natl. Acad. Sci. USA 2008, 105, 3374–3379. [Google Scholar] [CrossRef] [PubMed]
- Talia, M.; De Francesco, E.M.; Rigiracciolo, D.C.; Muoio, M.G.; Muglia, L.; Belfiore, A.; Maggiolini, M.; Sims, A.H.; Lappano, R. The G Protein-Coupled Estrogen Receptor (GPER) Expression Correlates with pro-Metastatic Pathways in ER-Negative Breast Cancer: A Bioinformatics Analysis. Cells 2020, 9, 622. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, J.; Ma, J.; Huang, J.; Wang, Y.; Wang, P.; Wang, F.; Tang, X. GPER Governs the Immune Infiltration of Gastric Cancer and Activates the NF-ΚB/ROS/Apoptosis Pathway in Gastric Mucosal Epithelium. Int. Immunopharmacol. 2023, 122, 110641. [Google Scholar] [CrossRef]
- Bertoni, A.P.S.; Manfroi, P.d.A.; Tomedi, J.; Assis-Brasil, B.M.; de Souza Meyer, E.L.; Furlanetto, T.W. The Gene Expression of GPER1 Is Low in Fresh Samples of Papillary Thyroid Carcinoma (PTC), and in Silico Analysis. Mol. Cell. Endocrinol. 2021, 535, 111397. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Zhan, N.; Li, R.; Dong, W. Downregulation of G Protein-Coupled Estrogen Receptor (GPER) Is Associated with Reduced Prognosis in Patients with Gastric Cancer. Med. Sci. Monit. 2019, 25, 3115–3126. [Google Scholar] [CrossRef]
- Gilligan, L.C.; Rahman, H.P.; Hewitt, A.M.; Sitch, A.J.; Gondal, A.; Arvaniti, A.; Taylor, A.E.; Read, M.L.; Morton, D.G.; Foster, P.A. Estrogen Activation by Steroid Sulfatase Increases Colorectal Cancer Proliferation via GPER. J. Clin. Endocrinol. Metab. 2017, 102, 4435–4447. [Google Scholar] [CrossRef]
- Bustos, V.; Nolan, Á.M.; Nijhuis, A.; Harvey, H.; Parker, A.; Poulsom, R.; Mcbryan, J.; Thomas, W.; Silver, A.; Harvey, B.J. GPER Mediates Differential Effects of Estrogen on Colon Cancer Cell Proliferation and Migration under Normoxic and Hypoxic Conditions. Oncotarget 2017, 8, 84258–84275. [Google Scholar] [CrossRef]
- Martin, S.G.; Lebot, M.N.; Sukkarn, B.; Ball, G.; Green, A.R.; Rakha, E.A.; Ellis, I.O.; Storr, S.J. Low Expression of G Protein-Coupled Oestrogen Receptor 1 (GPER) Is Associated with Adverse Survival of Breast Cancer Patients. Oncotarget 2018, 9, 25946–25956. [Google Scholar] [CrossRef] [PubMed]
- Samartzis, E.P.; Noske, A.; Meisel, A.; Varga, Z.; Fink, D.; Imesch, P. The G Protein-Coupled Estrogen Receptor (GPER) Is Expressed in Two Different Subcellular Localizations Reflecting Distinct Tumor Properties in Breast Cancer. PLoS ONE 2014, 9, e83296. [Google Scholar] [CrossRef]
- Filardo, E.J.; Graeber, C.T.; Quinn, J.A.; Resnick, M.B.; Giri, D.; DeLellis, R.A.; Steinhoff, M.M.; Sabo, E. Distribution of GPR30, a Seven Membrane-Spanning Estrogen Receptor, in Primary Breast Cancer and Its Association with Clinicopathologic Determinants of Tumor Progression. Clin. Cancer Res. 2006, 12, 6359–6366. [Google Scholar] [CrossRef] [PubMed]
- Sjöström, M.; Hartman, L.; Grabau, D.; Fornander, T.; Malmström, P.; Nordenskjöld, B.; Sgroi, D.C.; Skoog, L.; Stål, O.; Leeb-Lundberg, L.M.F.; et al. Lack of G Protein-Coupled Estrogen Receptor (GPER) in the Plasma Membrane Is Associated with Excellent Long-Term Prognosis in Breast Cancer. Breast Cancer Res. Treat. 2014, 145, 61–71. [Google Scholar] [CrossRef]
- Ignatov, A.; Ignatov, T.; Weißenborn, C.; Eggemann, H.; Bischoff, J.; Semczuk, A.; Roessner, A.; Costa, S.D.; Kalinski, T. G-Protein-Coupled Estrogen Receptor GPR30 and Tamoxifen Resistance in Breast Cancer. Breast Cancer Res. Treat. 2011, 128, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.; Liu, M.; Yang, F.; Luo, H.; Li, Z.; Tu, G.; Yang, G. GPR30 as an Initiator of Tamoxifen Resistance in Hormone-Dependent Breast Cancer. Breast Cancer Res. 2013, 15, R114. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhu, Q.; Liu, M.; Tu, G.; Li, Q.; Yuan, J.; Wen, S.; Yang, G. GPER Promotes Tamoxifen-Resistance in ER+ Breast Cancer Cells by Reduced Bim Proteins through MAPK/Erk-TRIM2 Signaling Axis. Int. J. Oncol. 2017, 51, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Chen, P.; Jiang, Q. G Protein-Coupled Estrogen Receptor Is a Critical Regulator in Metastasis of Breast Cancer Cells. J. Biosci. Med. 2017, 5, 127–140. [Google Scholar] [CrossRef][Green Version]
- Xu, T.; Ma, D.; Chen, S.; Tang, R.; Yang, J.; Meng, C.; Feng, Y.; Liu, L.; Wang, J.; Luo, H.; et al. High GPER Expression in Triple-Negative Breast Cancer Is Linked to pro-Metastatic Pathways and Predicts Poor Patient Outcomes. npj Breast Cancer 2022, 8, 100. [Google Scholar] [CrossRef]
- Chan, Y.; Lai, A.C.; Lin, R.; Wang, Y.; Wang, Y.; Chang, W.; Wu, H.; Lin, Y.; Chang, W.; Wu, J.; et al. GPER-Induced Signaling Is Essential for the Survival of Breast Cancer Stem Cells. Int. J. Cancer 2020, 1685, 1674–1685. [Google Scholar] [CrossRef] [PubMed]
- Albanito, L.; Madeo, A.; Lappano, R.; Vivacqua, A.; Rago, V.; Carpino, A.; Oprea, T.I.; Prossnitz, E.R.; Musti, A.M.; Andò, S.; et al. G Protein-Coupled Receptor 30 (GPR30) Mediates Gene Expression Changes and Growth Response to 17β-Estradiol and Selective GPR30 Ligand G-1 in Ovarian Cancer Cells. Cancer Res. 2007, 67, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, V.; Bauerschmitz, G.; Gallwas, J.; Gründker, C. Suppression of G Protein-Coupled Estrogen Receptor 1 (GPER1) Enhances the Anti-Invasive Efficacy of Selective ERβ Agonists. Anticancer Res. 2022, 42, 5187–5194. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Silva, C.D.; Riera-Leal, A.; Ortiz-Lazareno, P.C.; Jave-Suárez, L.F.; Ramírez De Arellano, A.; Lopez-Pulido, E.I.; Macías-Barragan, J.G.; Montoya-Buelna, M.; Dávila-Rodríguez, J.R.; Chabay, P.; et al. GPER Overexpression in Cervical Cancer Versus Premalignant Lesions: Its Activation Induces Different Forms of Cell Death. Anti-Cancer Agents Med. Chem. 2019, 19, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wu, Y.Z.; Zhang, Y.M.; Ji, X.H.; Hao, Q. Activation of G-Protein Coupled Estrogen Receptor Inhibits the Proliferation of Cervical Cancer Cells via Sustained Activation of ERK1/2. Cell Biochem. Funct. 2015, 33, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Akimoto, T.; Takasawa, A.; Takasawa, K.; Aoyama, T.; Murata, M.; Osanai, M.; Saito, T.; Sawada, N. Estrogen/GPR30 Signaling Contributes to the Malignant Potentials of ER-Negative Cervical Adenocarcinoma via Regulation of Claudin-1 Expression. Neoplasia 2018, 20, 1083–1093. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, Z.; Jiang, G.; Zhou, Y.; Yang, X.; Huang, H.; Liu, H.; Du, J.; Wang, H. Epigenetic down Regulation of G Protein-Coupled Estrogen Receptor (GPER) Functions as a Tumor Suppressor in Colorectal Cancer. Mol. Cancer 2017, 16, 87. [Google Scholar] [CrossRef]
- He, Y.Y.; Cai, B.; Yang, Y.X.; Liu, X.L.; Wan, X.P. Estrogenic G Protein-Coupled Receptor 30 Signaling Is Involved in Regulation of Endometrial Carcinoma by Promoting Proliferation, Invasion Potential, and Interleukin-6 Secretion via the MEK/ERK Mitogen-Activated Protein Kinase Pathway. Cancer Sci. 2009, 100, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.O.; Leslie, K.K.; Singh, M.; Qualls, C.R.; Revankar, C.M.; Joste, N.E.; Prossnitz, E.R. GPR30: A Novel Indicator of Poor Survival for Endometrial Carcinoma. Am. J. Obstet. Gynecol. 2007, 196, 386.e1–386.e11. [Google Scholar] [CrossRef]
- Petrie, W.K.; Dennis, M.K.; Hu, C.; Dai, D.; Arterburn, J.B.; Smith, H.O.; Hathaway, H.J.; Prossnitz, E.R. G Protein-Coupled Estrogen Receptor-Selective Ligands Modulate Endometrial Tumor Growth. Obstet. Gynecol. Int. 2013, 2013, 472720. [Google Scholar] [CrossRef]
- Skrzypczak, M.; Schüler, S.; Lattrich, C.; Ignatov, A.; Ortmann, O.; Treeck, O. G Protein-Coupled Estrogen Receptor (GPER) Expression in Endometrial Adenocarcinoma and Effect of Agonist G-1 on Growth of Endometrial Adenocarcinoma Cell Lines. Steroids 2013, 78, 1087–1091. [Google Scholar] [CrossRef]
- Lee, S.-J.; Kim, T.W.; Park, G.L.; Hwang, Y.S.; Cho, H.J.; Kim, J.-T.; Lee, H.G. G Protein-Coupled Estrogen Receptor-1 Agonist Induces Chemotherapeutic Effect via ER Stress Signaling in Gastric Cancer. BMB Rep. 2019, 52, 647–652. [Google Scholar] [CrossRef]
- Peña-Gutiérrez, K.M.; Hernández-Ortega, K.; Bello-Alvarez, C.; Camacho-Arroyo, I. Expression and Estrogen Regulation of G Protein–Coupled Estrogen Receptor in Human Glioblastoma Cells. Oncol. Lett. 2022, 24, 397. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Jiang, J.; Zheng, C.; Wang, N.; Zhai, X.; Fei, X.; Wu, R.; Jiang, X. Estrogen Nuclear Receptors Affect Cell Migration by Altering Sublocalization of AQP2 in Glioma Cell Lines. Cell Death Discov. 2018, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Chen, W.; Wen, L.; Zhang, J.; Zhang, Q.; Yang, J.; Liu, H.; Chen, B.W.; Zhou, Y.; Feng, X.; et al. G Protein-Coupled Estrogen Receptor Deficiency Accelerates Liver Tumorigenesis by Enhancing Inflammation and Fibrosis. Cancer Lett. 2016, 382, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Chaturantabut, S.; Shwartz, A.; Evason, K.J.; Cox, A.G.; Labella, K.; Schepers, A.G.; Yang, S.; Acuña, M.; Houvras, Y.; Mancio-Silva, L.; et al. Estrogen Activation of G-Protein–Coupled Estrogen Receptor 1 Regulates Phosphoinositide 3-Kinase and MTOR Signaling to Promote Liver Growth in Zebrafish and Proliferation of Human Hepatocytes. Gastroenterology 2019, 156, 1788–1804.e13. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.A.; Xiong, J.; Fu, Q.; Dong, Y.; Liu, M.; Peng, M.; Jin, W.; Zhou, L.; Xu, X.; Huang, X.; et al. GPER-Induced ERK Signaling Decreases Cell Viability of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 638171. [Google Scholar] [CrossRef]
- Ren, J.; Tao, Y.; Peng, M.; Xiao, Q.; Jing, Y.; Huang, J.; Yang, J.; Lin, C.; Sun, M.; Lei, L.; et al. Targeted Activation of GPER Enhances the Efficacy of Venetoclax by Boosting Leukemic Pyroptosis and CD8+ T Cell Immune Function in Acute Myeloid Leukemia. Cell Death Dis. 2022, 13, 915. [Google Scholar] [CrossRef]
- Torres-López, L.; Maycotte, P.; Liñán-Rico, A.; Liñán-Rico, L.; Donis-Maturano, L.; Delgado-Enciso, I.; Meza-Robles, C.; Vásquez-Jiménez, C.; Hernández-Cruz, A.; Dobrovinskaya, O. Tamoxifen Induces Toxicity, Causes Autophagy, and Partially Reverses Dexamethasone Resistance in Jurkat T Cells. J. Leukoc. Biol. 2019, 105, 983–998. [Google Scholar] [CrossRef]
- Lee, I.; Doepner, M.; Weissenrieder, J.; Majer, A.D.; Mercado, S.; Estell, A.; Natale, C.A.; Sung, P.J.; Foskett, J.K.; Carroll, M.P.; et al. LNS8801 Inhibits Acute Myeloid Leukemia by Inducing the Production of Reactive Oxygen Species and Activating the Endoplasmic Reticulum Stress Pathway. Cancer Res. Commun. 2023, 3, 1594–1606. [Google Scholar] [CrossRef]
- Liu, C.; Liao, Y.; Fan, S.; Fu, X.; Xiong, J.; Zhou, S.; Zou, M.; Wang, J. G-Protein-Coupled Estrogen Receptor Antagonist G15 Decreases Estrogen-Induced Development of Non-Small Cell Lung Cancer. Oncol. Res. 2019, 27, 283–292. [Google Scholar] [CrossRef]
- Li, Z.H.; Liu, C.; Liu, Q.H.; Wang, J.; Wang, Y.; Wang, Y.F.; Deng, S.J.; Li, D.B. Cytoplasmic Expression of G Protein-Coupled Estrogen Receptor 1 Correlates with Poor Postoperative Prognosis in Non-Small Cell Lung Cancer. J. Thorac. Dis. 2022, 14, 1466–1477. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yu, T.; Yang, F.; Han, J.; Zuo, B.; Huang, L.; Bai, X.; Jiang, M.; Wu, D.; Chen, S.; et al. G Protein-Coupled Estrogen Receptor Agonist G-1 Inhibits Mantle Cell Lymphoma Growth in Preclinical Models. Front. Oncol. 2021, 11, 668617. [Google Scholar] [CrossRef] [PubMed]
- Rudelius, M.; Rauert-Wunderlich, H.; Hartmann, E.; Hoster, E.; Dreyling, M.; Klapper, W.; Ott, G.; Rosenwald, A. The G Protein-Coupled Estrogen Receptor 1 (GPER-1) Contributes to the Proliferation and Survival of Mantle Cell Lymphoma Cells. Haematologica 2015, 100, e458–e461. [Google Scholar] [CrossRef]
- Fujiwara, S.; Terai, Y.; Kawaguchi, H.; Takai, M.; Yoo, S.; Tanaka, Y.; Tanaka, T.; Tsunetoh, S.; Sasaki, H.; Kanemura, M.; et al. GPR30 Regulates the EGFR-Akt Cascade and Predicts Lower Survival in Patients with Ovarian Cancer. J. Ovarian Res. 2012, 5, 35. [Google Scholar] [CrossRef]
- Smith, H.O.; Arias-Pulido, H.; Kuo, D.Y.; Howard, T.; Qualls, C.R.; Lee, S.J.; Verschraegen, C.F.; Hathaway, H.J.; Joste, N.E.; Prossnitz, E.R. GPR30 Predicts Poor Survival for Ovarian Cancer. Gynecol. Oncol. 2009, 114, 465–471. [Google Scholar] [CrossRef]
- Heublein, S.; Mayr, D.; Vrekoussis, T.; Friese, K.; Hofmann, S.S.; Jeschke, U.; Lenhard, M. The G-Protein Coupled Estrogen Receptor (GPER/GPR30) Is a Gonadotropin Receptor Dependent Positive Prognosticator in Ovarian Carcinoma Patients. PLoS ONE 2013, 8, e71791. [Google Scholar] [CrossRef]
- Heublein, S.; Mayr, D.; Friese, K.; Jarrin-Franco, M.C.; Lenhard, M.; Mayerhofer, A.; Jeschke, U. The G-Protein-Coupled Estrogen Receptor (GPER/GPR30) in Ovarian Granulosa Cell Tumors. Int. J. Mol. Sci. 2014, 15, 15161–15172. [Google Scholar] [CrossRef] [PubMed]
- Fraungruber, P.; Kaltofen, T.; Heublein, S.; Kuhn, C.; Mayr, D.; Burges, A.; Mahner, S.; Rathert, P.; Jeschke, U.; Trillsch, F. G Protein-Coupled Estrogen Receptor Correlates with Dkk2 Expression and Has Prognostic Impact in Ovarian Cancer Patients. Front. Endocrinol. 2021, 12, 564002. [Google Scholar] [CrossRef]
- Zhu, C.X.; Xiong, W.; Wang, M.L.; Yang, J.; Shi, H.J.; Chen, H.Q.; Niu, G. Nuclear G Protein-Coupled Oestrogen Receptor (GPR30) Predicts Poor Survival in Patients with Ovarian Cancer. J. Int. Med. Res. 2018, 46, 723–731. [Google Scholar] [CrossRef]
- Ignatov, T.; Modl, S.; Thulig, M.; Weißenborn, C.; Treeck, O.; Ortmann, O.; Zenclussen, A.C.; Costa, S.D.; Kalinski, T.; Ignatov, A. GPER-1 Acts as a Tumor Suppressor in Ovarian Cancer. J. Ovarian Res. 2013, 6, 51. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, H.; Wen, H.; Jiang, X.; Cao, X.; Zhang, G.; Liu, G. The Novel Estrogen Receptor GPER Regulates the Migration and Invasion of Ovarian Cancer Cells. Mol. Cell. Biochem. 2013, 378, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chan, Q.K.Y.; Lam, H.M.; Ng, C.F.; Lee, A.Y.Y.; Chan, E.S.Y.; Ng, H.K.; Ho, S.M.; Lau, K.M. Activation of GPR30 Inhibits the Growth of Prostate Cancer Cells through Sustained Activation of Erk1/2, c-Jun/c-Fos-Dependent Upregulation of P21, and Induction of G2 Cell-Cycle Arrest. Cell Death Differ. 2010, 17, 1511–1523. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.-Z.; Yan, R.-L.; Huang, J.-W.; Li, F.-L.; Zhong, Y.-X.; Chen, Y.; Liu, F.-N.; Hu, B.; Huang, S.-B.; Yin, L.-H. Activation of G Protein Coupled Estrogen Receptor (GPER) Promotes the Migration of Renal Cell Carcinoma via the PI3K/AKT/MMP-9 Signals. Cell Adhes. Migr. 2018, 12, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Yang, L.; Wang, N.; Li, L.; Xu, M.; Chen, G.G.; Liu, Z.-M. High Expression of GPER1, EGFR and CXCR1 Is Associated with Lymph Node Metastasis in Papillary Thyroid Carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 3213–3223. [Google Scholar] [PubMed]
- Ambrosini, G.; Natale, C.A.; Musi, E.; Garyantes, T.; Schwartz, G.K. The GPER Agonist LNS8801 Induces Mitotic Arrest and Apoptosis in Uveal Melanoma Cells. Cancer Res. Commun. 2023, 3, 540–547. [Google Scholar] [CrossRef]
- Cheng, S.-B.; Graeber, C.T.; Quinn, J.A.; Filardo, E.J. Retrograde Transport of the Transmembrane Estrogen Receptor, G-Protein-Coupled-Receptor-30 (GPR30/GPER) from the Plasma Membrane towards the Nucleus. Steroids 2011, 76, 892–896. [Google Scholar] [CrossRef]
- Prasad, V.V.T.S.; Gopalan, R.O.G. Continued Use of MDA-MB-435, a Melanoma Cell Line, as a Model for Human Breast Cancer, Even in Year, 2014. npj Breast Cancer 2015, 1, 15002. [Google Scholar] [CrossRef] [PubMed]
- Krakstad, C.; Trovik, J.; Wik, E.; Engelsen, I.B.; Werner, H.M.J.; Birkeland, E.; Raeder, M.B.; Øyan, A.M.; Stefansson, I.M.; Kalland, K.H.; et al. Loss of GPER Identifies New Targets for Therapy among a Subgroup of ERα-Positive Endometrial Cancer Patients with Poor Outcome. Br. J. Cancer 2012, 106, 1682–1688. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef]
- Notas, G.; Kampa, M.; Castanas, E. G Protein-Coupled Estrogen Receptor in Immune Cells and Its Role in Immune-Related Diseases. Front. Endocrinol. 2020, 11, 579420. [Google Scholar] [CrossRef] [PubMed]
- Di Vito, C.; Bergante, S.; Balduini, A.; Rastoldo, A.; Bagarotti, A.; Surico, N.; Bertoni, A.; Sinigaglia, F. The Oestrogen Receptor GPER Is Expressed in Human Haematopoietic Stem Cells but Not in Mature Megakaryocytes. Br. J. Haematol. 2010, 149, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Kolkova, Z.; Casslén, V.; Henic, E.; Ahmadi, S.; Ehinger, A.; Jirström, K.; Casslén, B. The G Protein-Coupled Estrogen Receptor 1 (GPER/GPR30) Does Not Predict Survival in Patients with Ovarian Cancer. J. Ovarian Res. 2012, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt Signaling in Cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, S.; Gu, L.; Di, W. Epigenetic Silencing of DKK2 and Wnt Signal Pathway Components in Human Ovarian Carcinoma. Carcinogenesis 2012, 33, 2334–2343. [Google Scholar] [CrossRef]
- Chen, K.; Chen, Z.; Wu, D.; Zhang, L.; Lin, X.; Su, J.; Rodriguez, B.; Xi, Y.; Xia, Z.; Chen, X.; et al. Broad H3K4me3 Is Associated with Increased Transcription Elongation and Enhancer Activity at Tumor-Suppressor Genes. Nat. Genet. 2015, 47, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Gao, Y.; Li, M.; Shi, J.; Peng, Y.; Du, X.; Klocker, H.; Sampson, N.; Shen, Y.; Liu, M.; et al. GPR30 Promotes Prostate Stromal Cell Activation via Suppression of Erα Expression and Its Downstream Signaling Pathway. Endocrinology 2016, 157, 3023–3035. [Google Scholar] [CrossRef] [PubMed]
- Madeo, A.; Maggiolini, M. Nuclear Alternate Estrogen Receptor GPR30 Mediates 17β-Estradiol–Induced Gene Expression and Migration in Breast Cancer–Associated Fibroblasts. Cancer Res. 2010, 70, 6036–6046. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, S.; Luo, H.; Chen, C.; Zhang, X.; He, L.; Tu, G. GPR30-Mediated HMGB1 Upregulation in CAFs Induces Autophagy and Tamoxifen Resistance in ERα-Positive Breast Cancer Cells. Aging 2021, 13, 16178–16197. [Google Scholar] [CrossRef]
- Zhang, R.; Zong, J.; Peng, Y.; Shi, J.; Du, X.; Liu, H.; Shen, Y.; Cao, J.; Jia, B.; Liu, F.; et al. GPR30 Knockdown Weakens the Capacity of CAF in Promoting Prostate Cancer Cell Invasion via Reducing Macrophage Infiltration and M2 Polarization. J. Cell. Biochem. 2021, 122, 1173–1191. [Google Scholar] [CrossRef]
- Yu, T.; Yang, G.; Hou, Y.; Tang, X.; Wu, C.; Wu, X.; Guo, L.; Zhu, Q.; Luo, H.; Du, Y.; et al. Cytoplasmic GPER Translocation in Cancer-Associated Fibroblasts Mediates CAMP/PKA/CREB/Glycolytic Axis to Confer Tumor Cells with Multidrug Resistance. Oncogene 2017, 36, 2131–2145. [Google Scholar] [CrossRef] [PubMed]
- Dumas, J.F.; Brisson, L. Interaction between Adipose Tissue and Cancer Cells: Role for Cancer Progression. Cancer Metastasis Rev. 2021, 40, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, L.; Yang, Z.; Tang, Y.; Tao, Y.; Zhan, Q.; Lei, L.; Jing, Y.; Jiang, X.; Jin, H.; et al. Glycolytic Enzyme PKM2 Mediates Autophagic Activation to Promote Cell Survival in NPM1-Mutated Leukemia. Int. J. Biol. Sci. 2019, 15, 882–894. [Google Scholar] [CrossRef]
- Rasoulpoor, S.; Asgharzadeh, M.R.; Shabani, S. A Mimic of the Tumor Microenvironment on GPR30 Gene Expression in Breast Cancer. Multidiscip. Cancer Investig. 2022, 6, 1–8. [Google Scholar] [CrossRef]
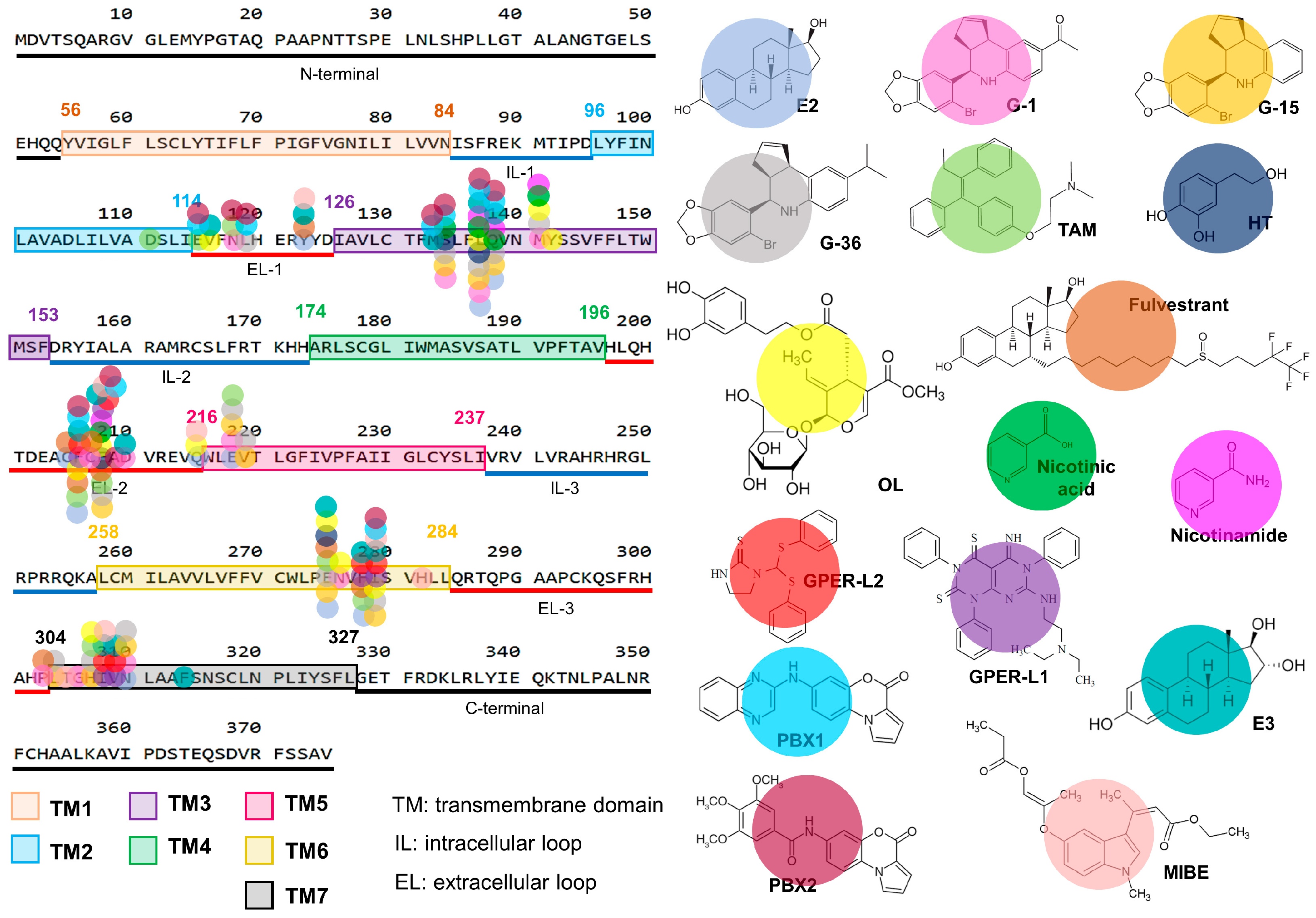
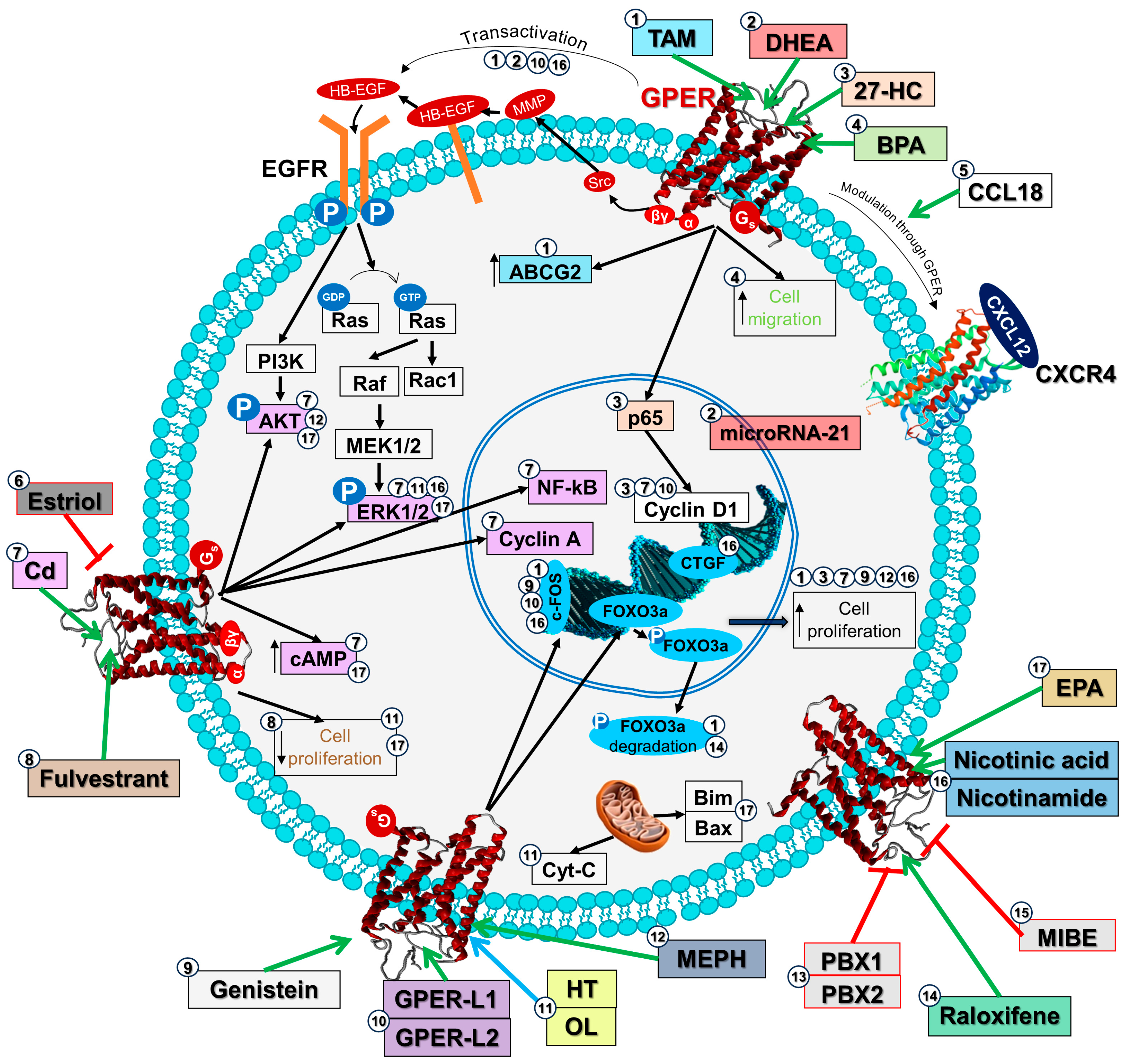


| Ligand | Model | Biological Effect | Reference |
|---|---|---|---|
| 4-OHTAM | MDA-MB-435 and HCC1806 (triple-negative breast cancer) cell lines | Increased cell numbers, caused transactivation of EGFR and c-fos expression through GPER a | [36] |
| WRO, FRO, and ARO (thyroid cancer) cell lines | Increased cell proliferation and c-fos expression through GPER a | [37] | |
| RL95-2 and HEC-1A (endometrial adenocarcinoma) cell lines | Increased cell growth, cyclin D1 expression, and EGFR transactivation through GPER a | [38] | |
| Ishikawa and RL95-2 (endometrial adenocarcinoma) cell lines | Induced cell migration and increased phosphorylated FAK levels through GPER a | [39] | |
| 27-HC | MDA-MB-231 (breast cancer) cell line | Increased cell proliferation and induced p65 nuclear translocation through GPER a,c | [40] |
| BPA | MDA-MB-231 (breast cancer) cell lines | Increased the migration of MDA-MB-231 cells through GPER a | [41] |
| Cadmium | WRO and FRO (thyroid cancer) cell lines | Induced cell proliferation, migration, invasion, ERK and AKT activation, nuclear NF-kB, secretion of IL-8, and cyclin A and D1 expression through GPER a | [42] |
| SKBR3 (breast cancer) cell line | Induced cell proliferation, stimulation of cAMP production, and ERK signaling pathway through GPER d | [43] | |
| CCL18 | Primary pre-B ALL samples; Nalm-6 (ALL) cell line | Through interaction with GPER b increased its expression Modulator of CXCR4-dependent responses | [44] |
| DHEA | HepG2 (hepatocellular carcinoma) cell line | Induced microRNA-21 transcription and EGFR transactivation through GPER a | [45] |
| EPA | ES2 and SKOV3 (ovarian cancer) cell lines | Decreased cell proliferation, induced cell apoptosis, increased p-AKT, p-ERK, pro-apoptotic Bim and Bax proteins, PKA activity, and cAMP expression through GPER a | [46] |
| ERα17p | MDA-MB-231 (breast cancer) cell lines | Decreased cell proliferation through GPER e,f | [47,48] |
| Estriol | SKBR3 (breast cancer) cell line | GPER c,e antagonist. Avoid GPER a activation (with G-1 and 4-OHTAM) effects: c-fos, p-ERKand CTGF expression, cell proliferation | [49] |
| Fulvestrant | MCF-7 (breast cancer) cell line | Decreased cell proliferation through GPER a | [50] |
| Genistein | WRO, FRO, and ARO (thyroid cancer) cell lines | Increased cell proliferation and c-fos expression through GPER a | [37] |
| GPER-L1 GPER-L2 | SKBR3 (breast cancer) and Ishikawa (endometrial cancer) cell lines | Increased cell growth, caused cyclin D1 protein induction and p-ERK and c-fos expression through GPER a,e | [51] |
| HT | SKBR3 (breast cancer) cell line | GPER c,e inverse agonist. Decreased cell viability, increased p-ERK1/2 levels and Cyt-c cytosolic release through GPER a. | [52] |
| MEHP | HeLa and SiHa (cervical cancer) cell lines | Increased cell proliferation and p-AKT and nuclear localization through GPER a | [53] |
| MIBE | SKBR3 (breast cancer) cell line | GPER c,e antagonist. Avoid GPER activation (with E2) effects: EGFR transactivation, cell proliferation, c-fos, and CTGF expression | [54] |
| Nicotinic acid Nicotinamide | SKBR3 (breast cancer) cell line and CAFs from breast cancer patients | Increased cell growth, migration, p-ERK1/2, c-fos and CTGF expression through GPER a,c,e in cells that do not express nicotinic acid receptor | [55] |
| OL | SKBR3 (breast cancer) cell line | GPER c,e inverse agonist. Decreased cell viability, increased p-ERK1/2 levels, and Cyt-c cytosolic release through GPER a. | [52] |
| PBX1 PBX2 | SKBR3 (breast cancer) cell line and CAFs from breast cancer patients | GPER c,e antagonist. Avoid GPER activation (with E2 and G-1) effects: cell growth, EGFR transactivation, p-ERK, c-fos, and CTGF expression | [56] |
| PLMI | MDA-MB-231 (breast cancer) cell lines | Decreased cell proliferation through GPER e,f | [47,48] |
| Raloxifene | MCF-7 (breast cancer) cell line | Induced FOXO3a inactivation through GPER a | [57] |
| TAM | MCF-7 (breast cancer) cell line | Induced FOXO3a inactivation through GPER a | [57] |
| MCF-7 (breast cancer) wild-type and TAM-resistant cell lines | Increased cell proliferation and caused transactivation of EGFR through GPER a Increased GPER translocation to the cell surface | [58] | |
| TAM-resistant MCF-7 (breast cancer) cell line | Caused ABCG2 upregulation, increased p-ERK and p-AKT levels, and caused EGFR transactivation through GPER a | [59] | |
| HEC-1A (endometrial adenocarcinoma) cell line | Increased cell proliferation through GPER a | [60] | |
| TAM-resistant MCF-7 (breast cancer) cell line | Induced β1-integrin expression through GPER a and EGFR | [61] |
| Type of Cancer | Model | Localization | Role in Cancer | Reference |
|---|---|---|---|---|
| Breast cancer (BC) | Tissue samples from early-stage invasive BC | Nucleus Cytoplasm | High nuclear GPER expression was associated with smaller tumors and a lower tumor grade Low cytoplasmic GPER expression was associated with adverse survival in BC patients treated with endocrine therapy | [102] |
| Primary invasive breast carcinoma samples Normal breast tissues T47D and MCF7 cell lines | Nucleus Cytoplasm | Cytoplasmic GPER expression was associated with better OS, a low tumor stage, and a luminal A/B subtype Nuclear GPER expression was associated with poorly differentiated carcinomas and triple-negative subtype | [103] | |
| Tissue samples from ductal carcinoma and a normal breast | Cytoplasm | GPER expression was associated with tumor size and metastases | [104] | |
| Tissue samples from tamoxifen-treated primary BC | Plasma membrane Cytoplasm Perinuclear | Plasma membrane GPER expression was associated with a poorer prognosis | [105] | |
| Tissue samples from tamoxifen-treated primary invasive BC or without tamoxifen treatment | Plasma membrane Cytoplasm Nucleus | GPER expression was associated with a shorter RFS in patients treated with tamoxifen and a favorable RFS in patients without tamoxifen treatment GPER expression is correlated with EGFR expression | [106] | |
| Tissue samples from tamoxifen-treated BC (metastatic BC and primary tumors) | Plasma membrane Cytoplasm | GPER expression is correlated with EGFR expression Higher GPER expression in metastatic cancer compared to primary tumor | [107] | |
| Tissue samples from recurred BC and primary tumors | Cytoplasm | Higher GPER expression in recurred tumors compared to primary tumors, where GPER expression had an inverse correlation with pro-apoptotic Bim protein | [108] | |
| Tissue samples from BC | Cytoplasm | GPER and CXCR1 expression in advanced stages of BC. | [109] | |
| Tissue samples from triple-negative BC | Cytoplasm | Higher GPER expression is correlated with necrosis in the cancer nest, increased metastatic events, mesenchymal-like subtypes, and worse DMFS | [110] | |
| Tissue samples from tamoxifen-treated BC (metastatic BC and primary tumors) Tamoxifen resistant MCF-7 cell line | N.D. | Higher GPER expression in metastatic cancer compared to primary tumors. GPR30 expression is correlated with ABCG2 expression GPER activation * (with G-1) caused ABCG2 upregulation, increased p-ERK and p-AKT levels, and caused EGFR transactivation | [59] | |
| BC stem cells and non-BC stem cells from patient-derived xenografts | N.D. | Higher GPER expression in BC stem cells compared to non-BC stem cells GPER silencing reduced cell growth in BC stem cells | [111] | |
| SKBR3 cell line | N.D. | GPER activation * (with G-1, E2) caused cell proliferation | [112] | |
| SKBR3 and BT-20 cell lines | N.D. | GPER activation * (with E2) increased p-ERK and p-AKT levels, migration, and IL-8 secretion | [76] | |
| MDA-MB-435 and HCC1806 cell lines | N.D. | GPER activation * (with E2) increased cell number, c-fos expression, and EGFR transactivation | [36] | |
| SKBR3 and MDA-MB-231 cell lines | N.D. | GPER activation * (with G-1) decreased cell viability | [6] | |
| MDA-MB-435 and MDA-MB-231 cell lines | N.D. | GPER activation * (with G-1) decreased cell viability, caused cell death, and caused cell cycle arrest | [7] | |
| MCF-7 cell line | N.D. | GPER activation * (with E2, G-1) induced FOXO3a inactivation | [57] | |
| MCF-7, MDA-MB-231, and Bcap-37 cell lines | N.D. | GPER inhibition * (or G-15 treatment) increased doxorubicin sensitivity and avoid doxorubicin-induced EMT | [31] | |
| MDA-MB-231 and HCC1806 cell lines | N.D. | GPER inhibition * decreased cell invasion and sensitized cells to tamoxifen | [113] | |
| SKBR3 cell line and CAFs | N.D. | GPER activation * (with G-1, E2) increased c-fos and FASN expression through EGFR transactivation | [77] | |
| SKBR3 cell line | N.D. | GPER activation ** (with E2) induced cell proliferation, stimulation of cAMP production, and the ERK signaling pathway | [43] | |
| SKBR3 cell line and CAFs | N.D. | GPER activation * (with G-1, E2) increased cell proliferation, c-fos expression, and downregulated microRNA-338-3p | [78] | |
| Tissue samples from CC tumors, healthy tissue, and precursor lesions | Cytoplasm Nucleus | Higher cytoplasmic and nuclear GPER expression in cervical cancer compared to healthy tissue | [114] | |
| Cervical cancer (CC) | HeLa, SiHa, C-33A and CaSki cell lines Tissue samples from normal cervical epithelial cells | N.D. | GPER activation * (with G-1) decreased cell viability, caused cell cycle arrest, and caused EGFR transactivation | [115] |
| HeLa, SiHa, and C-33A cell lines | N.D. | GPER inhibition * increased stem cell properties and invasive behavior; enhanced EMT | [91] | |
| Surgical specimens from cervical adenocarcinoma patients HCA1 cell line | N.D. | Co-expression of GPER and claudin-1 is correlated with a poor prognosis GPER activation * (with G-1) increased claudin-1 expression | [116] | |
| Colorectal cancer (CRC) | Tissue samples from CRC tumors and adjacent normal tissues | N.D. | Lower GPER expression in CRC compared to normal tissues Low GPER expression was associated with a poorer survival rate | [117] |
| HT-29, DLD-1, and HCT116 cell lines | N.D. | GPER activation * (with G-1, E2) decreased ATM; under normoxic conditions, decreased cell proliferation, migration, VEGFA, and HIF1-α expression; and increased them in a hypoxic environment | [101] | |
| LoVo cell line | N.D. | GPER activation * (with G-1, E2) increased c-fos and FASN expression through EGFR transactivation | [77] | |
| HCT116 cell line Tissue samples from CRC tumors and matched normal tissues | N.D. | GPER activation * (with G-1, E2) induced cell proliferation GPER expression is correlated with CTGF expression | [100] | |
| Endometrial cancer (EC) | Endometrial tissues from patients with BC treated or not with tamoxifen | Nucleus Cytoplasm | GPER expression is correlated with the time of tamoxifen treatment and the development of an endometrial abnormality Mostly expressed in abnormal endometrial tissue of patients treated with tamoxifen | [60] |
| Endometrial tissues from patients with BC treated or not with tamoxifen | Plasma membrane Cytoplasm | Higher GPER in expression EC tissue of patients treated with tamoxifen, which had a worse prognosis | [38] | |
| Tissue samples from uterine EC KLE and RL95-2 cell lines | Plasma membrane Cytoplasm | Higher GPER expression in uterine EC compared to normal tissues GPER inhibition * decreased cell growth. GPER activation * (with E2, G-1) increased cell viability | [118] | |
| Tissue samples from the uterine EC | Subcellular | GPER expression correlated with EGFR expression but negatively with PR GPER overexpression was associated with poorer survival | [119] | |
| Hec50 cell line | Cytoplasm | GPER activation * (with E2)-induced PI3K activation | [120] | |
| Tissue samples from EC tumors Ishikawa and RL95-2 cell lines | N.D. | GPER expression is correlated with pFAK expression GPER activation * (with E2 and G-1) induced cell migration and increased pFAK levels | [39] | |
| Tissue samples from endometrial adenocarcinoma tumors and normal endometrium HEC-1A, HEC-1B, and RL95-2 cell lines | N.D. | Lower GPER expression in endometrial adenocarcinoma compared to normal tissues. GPER expression was associated with PR and PTEN expression G-1 decreased cell viability without effect on the GPER-negative cell line | [121] | |
| Gastric cancer (GC) | Tissue samples from GC tumors and adjacent normal gastric HGC-27, MGC-803, SGC-7901, and GES-1 cell lines | Nucleus Cytoplasm | Lower GPER expression in GC compared to normal tissues and cells Low GPER mRNA levels predict a poor prognosis | [99] |
| Tissue samples from GC tumors and normal gastric AGS, SNU-216, NCI-N87, SNU-620, SNU-638, SNU-668, NUGC-3, and MKN-74 cell lines | N.D. | Lower GPER expression in GC compared to normal tissues GPER activation * (with G-1) in highly GPER-expressive cell lines decreases cell viability. Without effect in the GPER-low cell line | [122] | |
| AGS and BGC-823 cell lines | N.D. | GPER induces cisplatin resistance by promoting EMT GPER activation * (with G-1) reduced cisplatin sensitivity. GPER inhibition * (or G-15 treatment) increased cisplatin sensitivity | [32] | |
| AGS and MGC-803 cell lines | N.D. | GPER inhibition * decreased cell viability, proliferation, migration, invasion, EMT, cyclin D1, CDK4, p-PI3K, p-AKT, p-mTOR, MMP2, and MMP9 levels. Contrary effects with GPER overexpression | [88] | |
| Glioma | U87, LN229, T98, and U251 cell lines | Cytoplasm Plasma membrane | Lower GPER (only mRNA) expression between glioma tissue and human astrocytes | [123] |
| LN229 and U251 cell lines | Plasma membrane Cytoplasm Around the nucleus | N.D. | [29] | |
| U87 and T98G cell lines | N.D. | Without differences in GPER expression between glioma tissue and glial cells | [124] | |
| Hepatocellular carcinoma (HCC) | Tissue samples from HCC tumors and adjacent normal tissues | Cytoplasm Plasma membrane | Lower GPER expression in HCC compared to normal tissues | [125] |
| Tissue samples from HCC tumors, cirrhotic livers and normal liver | Cytoplasm | Higher GPER expression in HCC and cirrhotic livers compared to normal liver | [126] | |
| Tissue samples from HCC tumors and paired normal tissues HCCLM3 cell line | Cytoplasm | Lower GPER expression in HCC compared to normal tissues. GPER expression is correlated with better OS. Co-expression of GPER and p-ERK predicted an improved prognosis GPER activation * (with G-1) increased p-ERK levels and caused cell cycle arrest and apoptosis | [127] | |
| HepG2 cell line | N.D. | GPER activation * (with G-1, E2) increased c-fos and FASN expression through EGFR transactivation | [77] | |
| HepG2 cell line | N.D. | GPER activation * (with G-1) induced microRNA-21 transcription | [45] | |
| Leukemia | Primary pre-B ALL samples; Nalm-6 cell line | Plasma membrane | Expressed in primary cells from pre-B ALL but not from common ALL | [44] |
| Primary AML samples and bone marrow mononuclear cells from healthy donors NB4, U937, KG1a, THP-1, and OCI-AML2 cell lines | Plasma membrane | Low GPER expression in B-ALL blast compared to bone marrow mononuclear cells from healthy donors GPER activation * (with G-1) induced apoptosis and enhanced the efficacy of venetoclax | [128] | |
| Jurkat, CCRF-CEM, and Molt-3 cell lines | Plasma membrane | Low GPER expression in T-ALL blasts compared to CD4+ lymphocytes | [30] [129] | |
| HL-60 cell line | Plasma membrane Cytoplasm | N.D. | [34] | |
| Primary AML samples: MOLM14, U937, HL60, and THP-1 cell lines | N.D. | N.D. | [130] | |
| Lung cancer (LC) | Tissue samples from NSCLC and benign pulmonary lesions | Cytoplasm Nucleus | Higher GPER expression in NSCLC compared to benign pulmonary tissue Higher cytoplasmic GPER expression was associated with higher stages and poorer differentiation. Cytoplasmic GPER is correlated with ER-β | [131] |
| Tissue samples from NSCLC | Cytoplasm Nucleus | Co-expression of cytoplasmic and nuclear GPER expression was associated with a poor prognosis. Nuclear GPER is correlated with ER-β | [132] | |
| Tissue samples from LC tumors and adjacent normal tissues A549, H23, H1299, H1792, H1395, H1435, H1793, H1944, H2073 (LC), and HPL1D, HBEC2-E, HBEC2-KT, and HBEC3-KT (epithelial lung) cell lines | N.D. | Higher GPER expression in LC compared to normal tissues Higher GPER expression in LC cell lines compared to epithelial lung cell lines | [13] | |
| A549 and H1299 cell lines | N.D. | GPER inhibition * decreased cell growth, NOTCH1, HIF-1α, and CXCR4 mRNA levels; Contrary effects with GPER overexpression | [92] | |
| Mantle cell lymphoma (MCL) | MCL samples Jeko-1, Mino, Rec-1, and Granta-519 cell lines | Nucleus | GPER activation * (with G-1) caused cell death and enhanced the antiproliferative effect of ibrutinib GPER inhibition * increased cell proliferation | [133] |
| MCL samples Rec-1, Granta-519, Mino, Jeko-1, Hbl-2, Nceb-1, Upn-1, Upn-2, L128 and Z138C cell lines | Nucleus | GPER inhibition * decreased p-p42/44 (ERK1/2), p-AKT, and cyclin D1 levels. GPER inhibition (with G-36) reduced cell proliferation and potentiated the effects of paclitaxel. Cell lines with low GPER expression did not respond to G-36 | [134] | |
| Ovarian cancer (OC) | Primary epithelial OC samples Low malignant potential tumors | Cytoplasm | Higher GPER expression in OC compared to borderline malignancies Co-expression of GPER-EGFR was associated with a poorer PFS | [135] |
| Epithelial OC samples Low malignant potential tumors | Nucleus | Greater frequency of high GPER in OC compared to low-malignant ovarian tumors. GPER is preferably expressed in high-risk OC. High GPER expression was associated with poorer survival | [136] | |
| Epithelial OC samples | Cytoplasm | GPER expression is correlated with FSHR and LHCGR. High GPER was associated with better OS in FSHR/LHCGR negative patients. Higher GPER expression in well-differentiated carcinomas compared to poorly differentiated carcinomas | [137] | |
| Ovarian granulosa cell tumor samples | Cytoplasm | High GPER was associated with a poorer outcome in newly-diagnosed ovarian granulosa cell tumors | [138] | |
| Epithelial OC samples | Cytoplasm | Cytoplasmic GPER is correlated with Dkk2 expression. Patients with high co-expression of GPER and Dkk2 had better OS | [139] | |
| Primary epithelial OC samples SKOV3 cell line | Cytoplasm Nucleus | Nuclear GPER expression was an independent negative prognostic factor for OS | [140] | |
| Epithelial OC samples Caov3 and Caov4 cell lines | Cytoplasm Plasma membrane | Co-expression with H3K4me3 correlates with a favorable prognosis GPER activation * (with G-1) attenuated cell proliferation/migration and increased H3K4me3 and p-ERK1/2 levels | [35] | |
| OC, low-malignant tumors, and benign ovarian tumors samples SKOV3 and OVCAR3 cell lines | N.D. | Lower GPER expression in OC compared to benign and low-malignant ovarian tumors. GPER expression was associated with favorable DFS | [141] | |
| OVCAR5 cell line | N.D. | GPER activation * (with E2, G-1) increased cell migration, invasion, MMP-9 expression, and activity. Contrary effects with GPER inhibition * | [142] | |
| BG-1 and 2008 cell lines | N.D. | GPER activation * (with G-1, E2) causes cell proliferation | [112] | |
| SKOV3 cell line | N.D. | GPER inhibition * decreased cell number, migration, invasion, c-fos, and cyclin D1 expression | [79] | |
| Prostate cancer | PC-3 cell line | N.D. | GPER activation * (with G-1) decreased cell growth, increased p-ERK and p21 levels | [143] |
| Renal cell carcinoma | ACHN, OS-RC-2, and SW839 cell lines | N.D. | GPER activation * (with G-1) increased cell migration and invasion | [144] |
| Thyroid cancer | Tissues samples from papillary thyroid carcinoma (PTC), nodular hyperplasia, and normal thyroid | Cytoplasm Plasma membrane | GPER expression correlated with EGFR and CXCR1 expression; expression of these receptors correlated with lymph node metastasis. Higher GPER, EGFR, and CXCR1 expression in PTC compared to nodular hyperplasia and normal thyroid tissues | [145] |
| Tissue samples from malignant follicular thyroid carcinoma (FTC), benign follicular thyroid adenoma (FTA), and normal thyroid | Cytoplasm Plasma membrane | Higher GPER expression in malignant FTC compared to benign FTA, without significant differences between FTA and normal thyroid tissues. GPER expression is correlated with EGFR and CXCR1 expression in malignant FTC | [87] | |
| Tissues samples from PTC, follicular variant of PTC, and adjacent non-malignant tissues | Cytoplasm | Lower GPER (mRNA) expression in PTC compared to non-malignant tissues, without differences in protein expression. Lower GPER (mRNA) expression in classical PTC compared to the follicular variant of PTC | [98] | |
| Uveal melanoma | Omm1.3 and 92.1 cell lines | N.D. | GPER inhibition * induced cell growth and downregulation of p53 | [146] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-López, L.; Olivas-Aguirre, M.; Dobrovinskaya, O. The G Protein-Coupled Estrogen Receptor GPER in the Development and Progression of Cancer. Receptors 2024, 3, 220-254. https://doi.org/10.3390/receptors3020012
Torres-López L, Olivas-Aguirre M, Dobrovinskaya O. The G Protein-Coupled Estrogen Receptor GPER in the Development and Progression of Cancer. Receptors. 2024; 3(2):220-254. https://doi.org/10.3390/receptors3020012
Chicago/Turabian StyleTorres-López, Liliana, Miguel Olivas-Aguirre, and Oxana Dobrovinskaya. 2024. "The G Protein-Coupled Estrogen Receptor GPER in the Development and Progression of Cancer" Receptors 3, no. 2: 220-254. https://doi.org/10.3390/receptors3020012
APA StyleTorres-López, L., Olivas-Aguirre, M., & Dobrovinskaya, O. (2024). The G Protein-Coupled Estrogen Receptor GPER in the Development and Progression of Cancer. Receptors, 3(2), 220-254. https://doi.org/10.3390/receptors3020012







