Near Real-Time Detection of E. coli in Reclaimed Water
Abstract
:1. Introduction
2. Methods
2.1. Multi-Angle Light Scattering Technology (Sensor A)
2.2. Real-Time Detection via Fluorescence Emission (Sensor B)
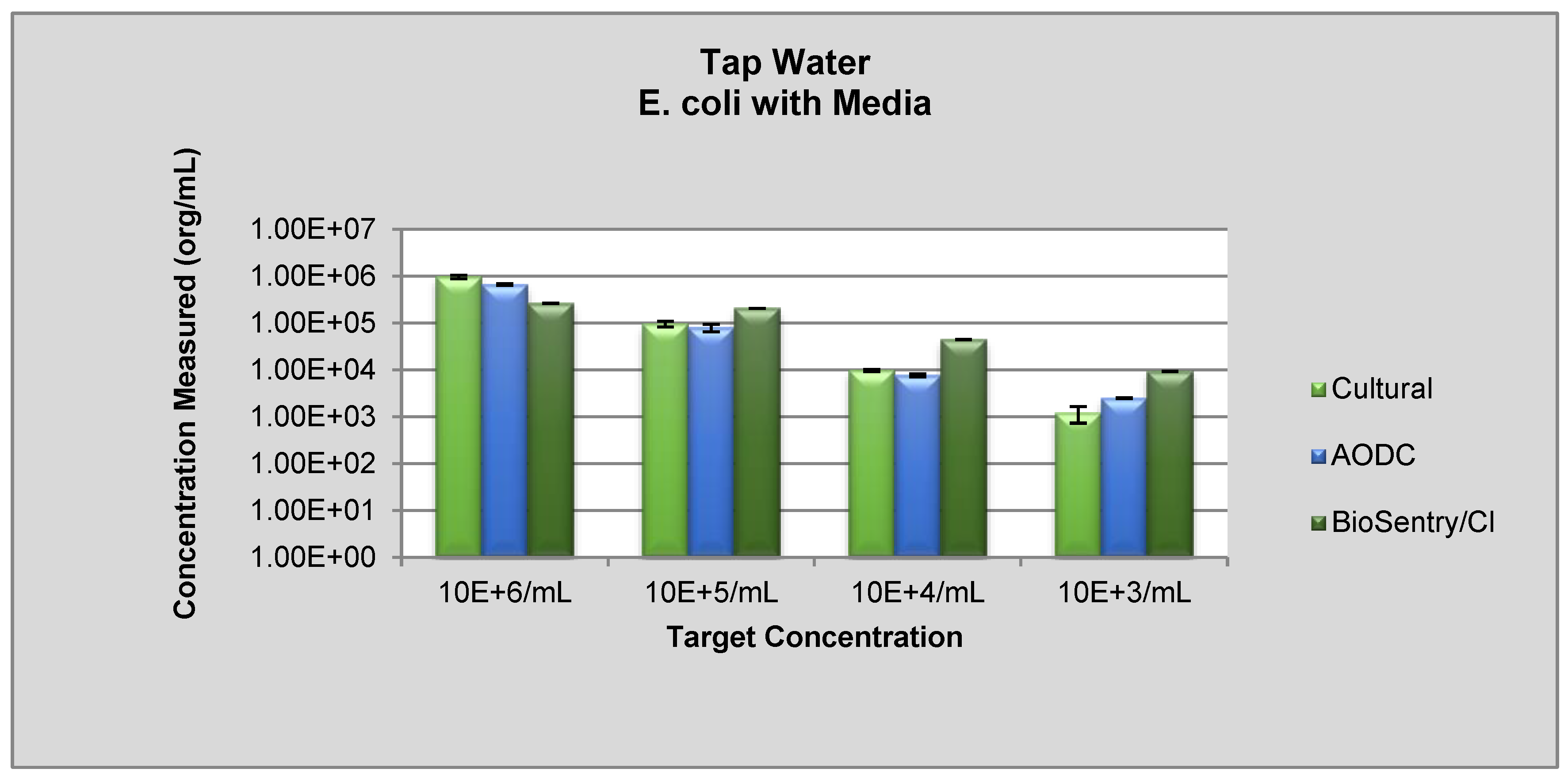
2.2.1. Real-Time Monitoring of E. coli in Distilled and Tap Water
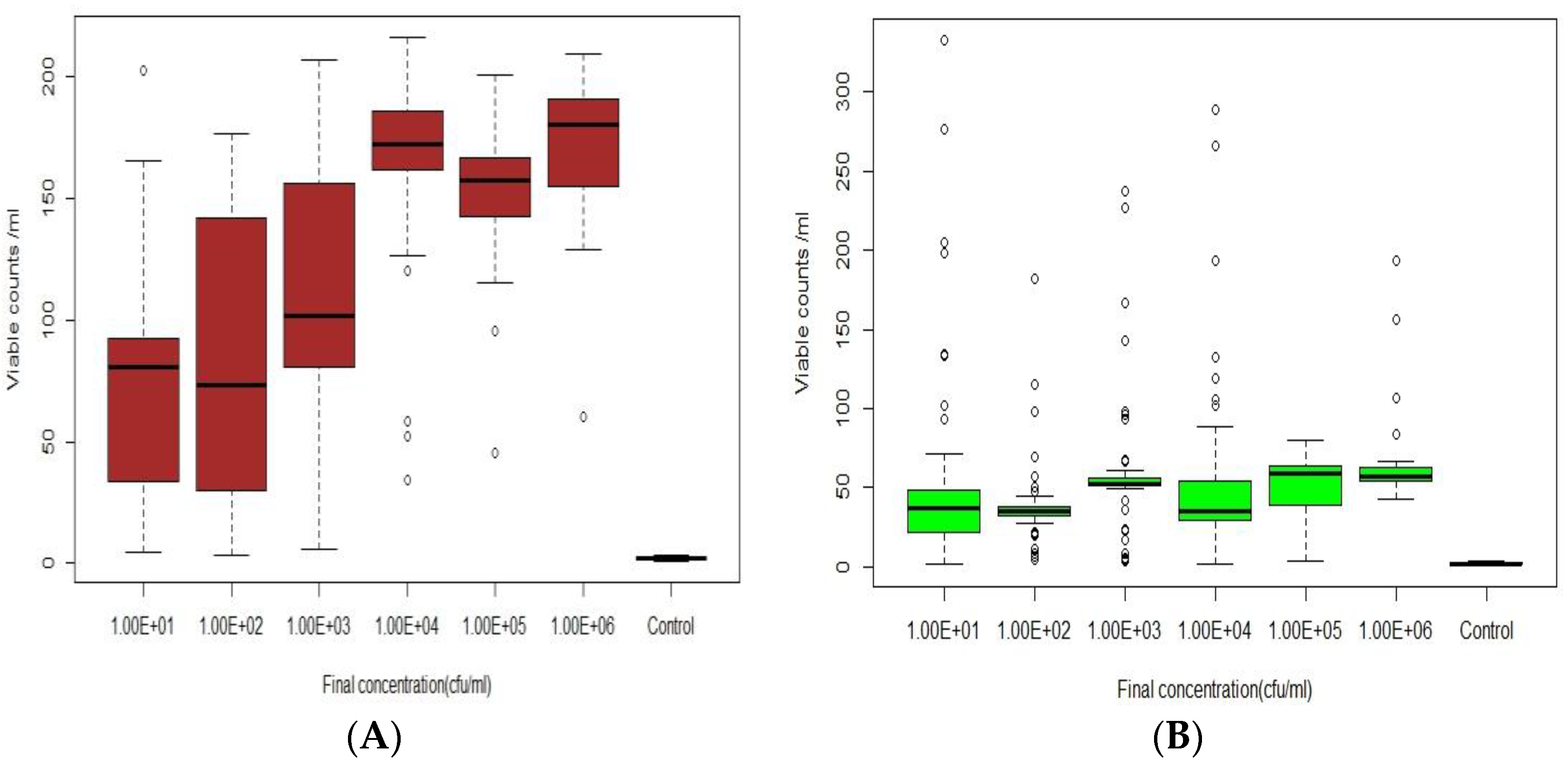
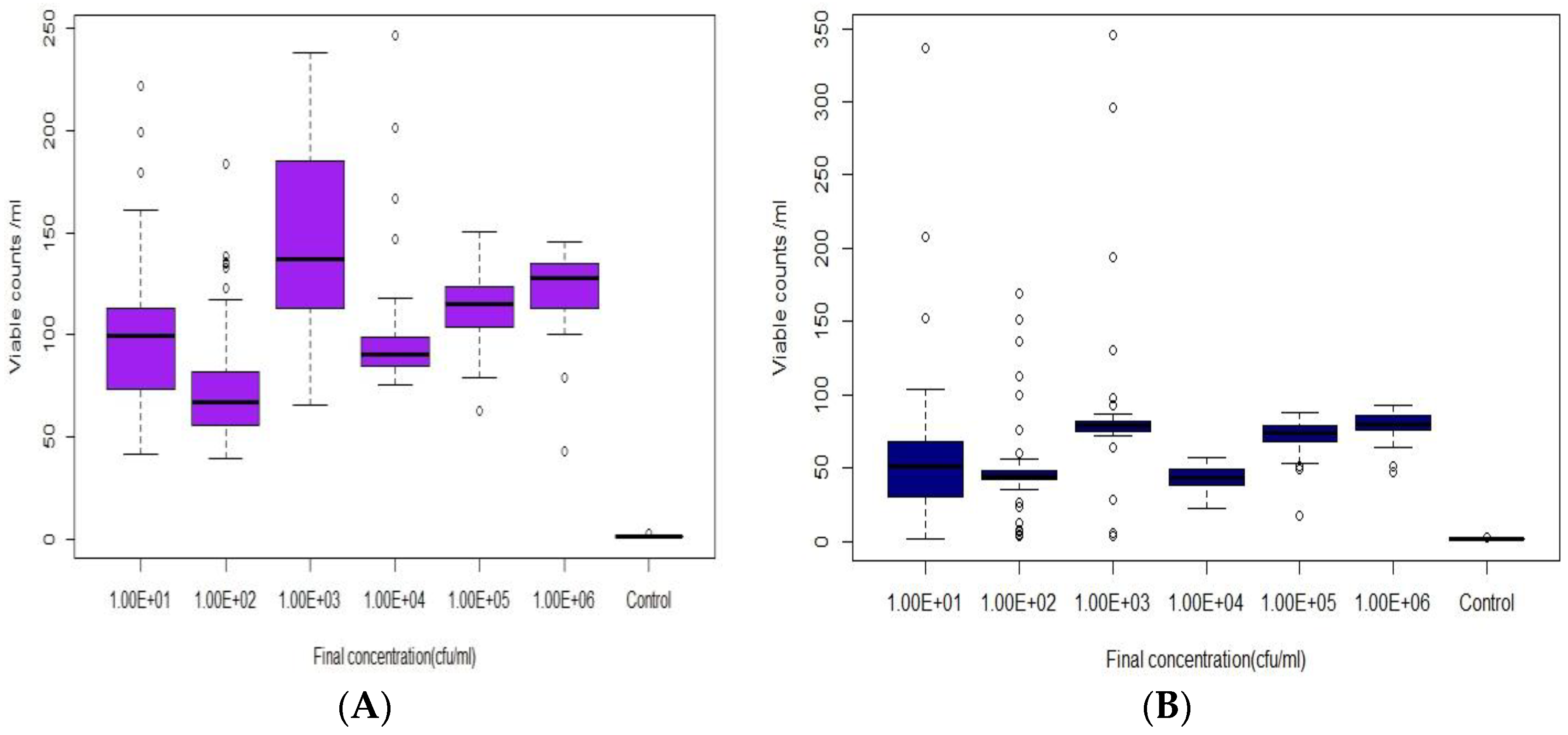
2.2.2. Real-Time Detection of E. coli in Modelled Reclaimed Water
2.3. Real-Time Detection via ATP Production (Sensor C)
2.4. Data Analysis
3. Results and Discussion
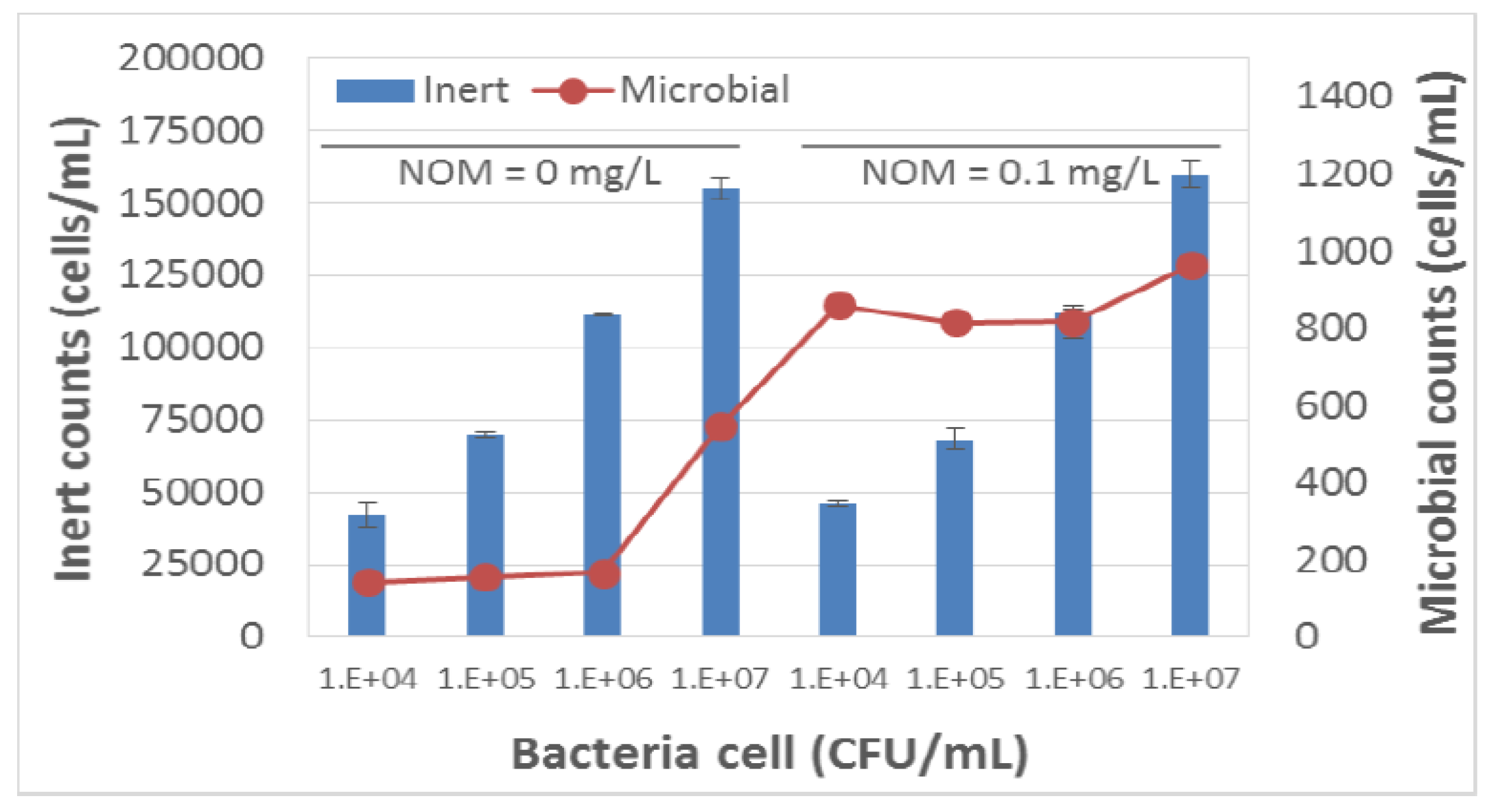
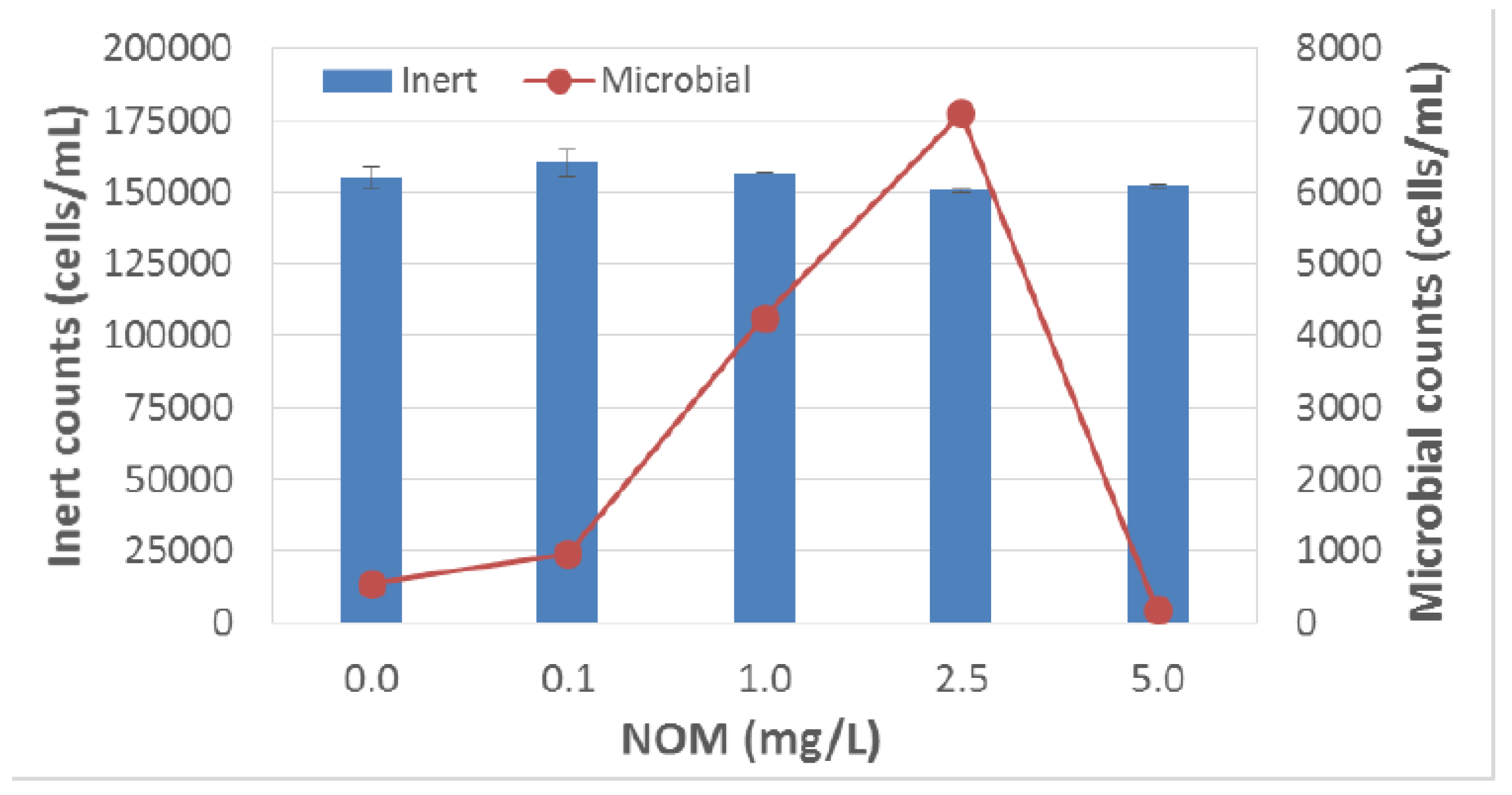
3.1. Multi-Angle Light Scattering Technology [Sensor A]
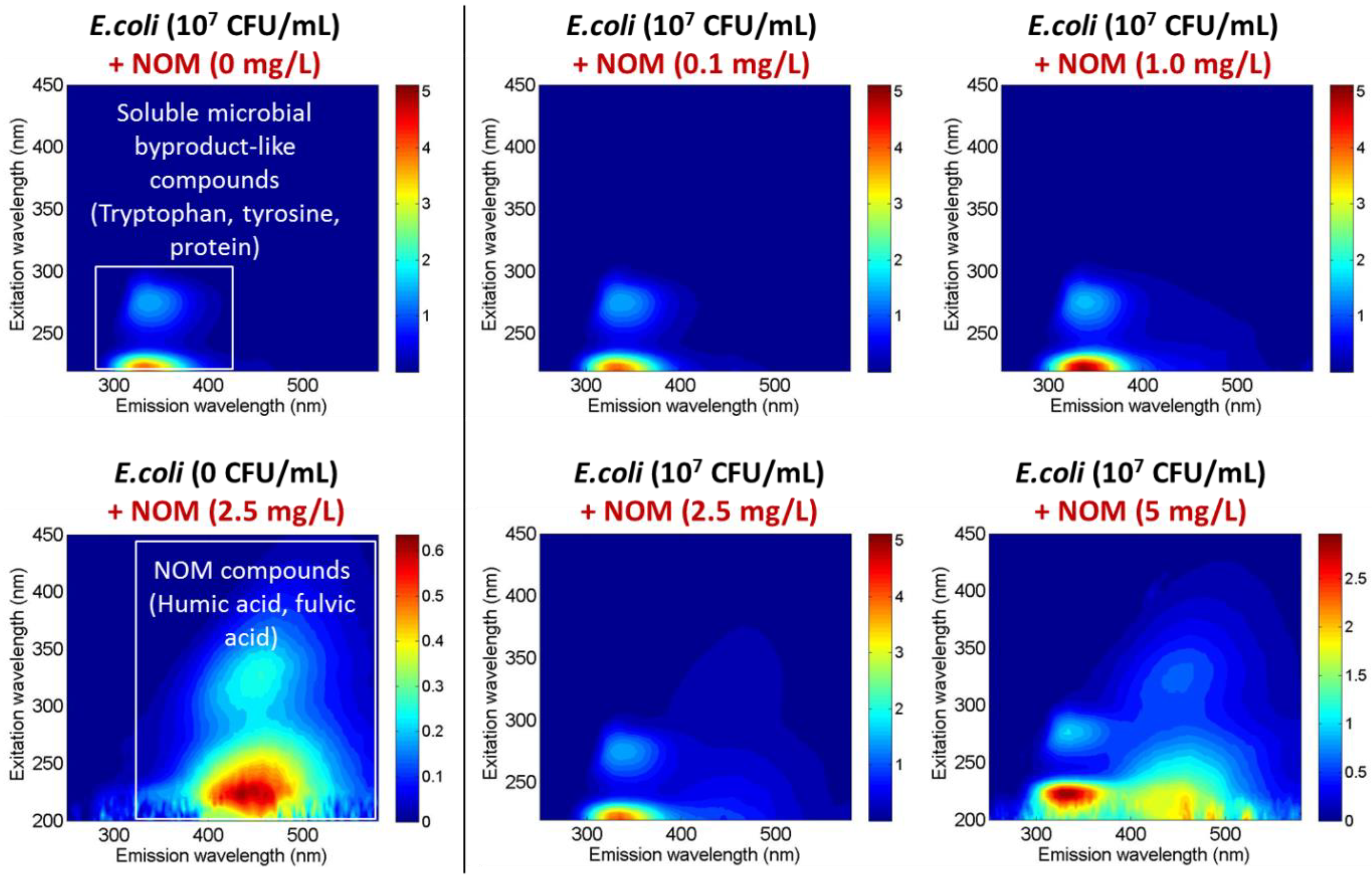
3.2. Real-Time Detection via Fluorescence Emission [Sensor B]
3.3. Real-Time Detection via ATP Production [Sensor C]
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Scott, B.A.; Pepper, I.L. Water distribution systems as living ecosystems: Impact on taste and odor. J. Environ. Sci. Health Part A 2010, 45, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Hijnem, W.A.M.; Beerendonk, E.F.; Medema, G.J. Inactivation credit of UV radiation for viruses, bacteria and protozoan cysts in water: A review. Water Res. 2006, 40, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Roszack, D.B.; Collwell, R.R. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 1987, 51, 365–378. [Google Scholar]
- Wols, B.A.; Harmsen, D.J.H.; Beerendonk, E.F.; Hofman-Caris, C.H.M. Predicting pharmaceutical degradation by UV[MP] H2O2 processes: A kinetic model. Chem. Eng. J. 2015, 263, 336–345. [Google Scholar] [CrossRef]
- Zaleski, K.J.; Josephson, K.L.; Gerba, C.P.; Pepper, I.L. Survival, growth and regrowth of enteric indicator and pathogenic bacteria in biosolids, compost, soil and land applied biosolids. J. Res. Sci. Technol. 2005, 2, 49–63. [Google Scholar]
- Sherchan, S.; Gerba, C.; Pepper, I. Evaluation of real-time water quality sensors for the detection of intentional bacterial spore contamination of potable water. J. Biosens. Bioelectron. 2013, 4, 141–145. [Google Scholar]
- Chen, W.; Westerhoff, P.; Leenheer, J.A.; Booksh, K. Fluorescence Excitation-Emission Matrix Regional Integration to Quantify Spectra for Dissolved Organic Matter. Environ. Sci. Technol. 2003, 37, 5701–5710. [Google Scholar] [CrossRef] [PubMed]
- Sherchan, S.; Snyder, S.; Gerba, G.; Pepper, I. Inactivation of MS2 coliphage by UV and hydrogen peroxide: Comparison by cultural and molecular methodologies. J. Environ. Sci. Health A 2014, 49, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Pepper, I.L.; Sherchan, S.P.; Miles, S.L.; Clarke, B.O.; Snyder, S.A. Ensuring safe water through advanced oxidation and real-time sensors. In Proceedings of the WDSA 2012: 14th Water Distribution Systems Analysis Conference, Adelaide, South Australia, 24–27 September 2012. [Google Scholar]
- Storey, M.V.; Gaag, B.V.; Burns, B.P. Advances in on-line drinking water quality monitoring and early warning systems. Water Res. 2011, 45, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Che, H.; Smith, K.; Chang, T. A real time method of contaminant classification using conventional water quality sensors. J. Environ. Manag. 2015, 154, 13–21. [Google Scholar] [CrossRef] [PubMed]

| Sensor C (ME/100 mL) | HPC (Counts/100 mL) | ||
|---|---|---|---|
| GV 1 | Feed water | 4.4 × 107 | 3.8 × 108 |
| Permeate | 2.4 × 106 | 2.6 × 106 | |
| Brine | 2.4 × 108 | 2.2 × 108 | |
| Ina 2 | Feed water | 9.2 × 107 | 2.6 × 108 |
| Permeate | 6.2 × 106 | 1.0 × 107 | |
| Brine | 4.1 × 108 | 2.9 × 108 |
| Target Blending Ratio (%Brine:%Permeate) | Salt Passage (%) | Sensor C (ME/100 mL) | HPC (Counts/100 mL) | |
|---|---|---|---|---|
| GV | 0.1 | 0.2 | 3.0 | 2.8 |
| 0.5 | 2.0 | 3.1 | 2.5 | |
| 1 | 4.6 | 3.0 | 2.5 | |
| 2 | 9.1 | 6.9 | 2.1 | |
| Ina | 0.1 | 0.6 | 5.6 | 12.5 |
| 0.5 | 2.3 | 6.3 | 14.5 | |
| 1 | 4.7 | 6.1 | 16.0 | |
| 2 | 9.3 | 10.1 | 18.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sherchan, S.; Miles, S.; Ikner, L.; Yu, H.-W.; Snyder, S.A.; Pepper, I.L. Near Real-Time Detection of E. coli in Reclaimed Water. Sensors 2018, 18, 2303. https://doi.org/10.3390/s18072303
Sherchan S, Miles S, Ikner L, Yu H-W, Snyder SA, Pepper IL. Near Real-Time Detection of E. coli in Reclaimed Water. Sensors. 2018; 18(7):2303. https://doi.org/10.3390/s18072303
Chicago/Turabian StyleSherchan, Samendra, Syreeta Miles, Luisa Ikner, Hye-Weon Yu, Shane A. Snyder, and Ian L. Pepper. 2018. "Near Real-Time Detection of E. coli in Reclaimed Water" Sensors 18, no. 7: 2303. https://doi.org/10.3390/s18072303
APA StyleSherchan, S., Miles, S., Ikner, L., Yu, H.-W., Snyder, S. A., & Pepper, I. L. (2018). Near Real-Time Detection of E. coli in Reclaimed Water. Sensors, 18(7), 2303. https://doi.org/10.3390/s18072303






