B Chromosomes in the Drosophila Genus
Abstract
:1. Introduction
2. B Chromosomes within the Genus Drosophila
3. B Chromosomes in Drosophila melanogaster
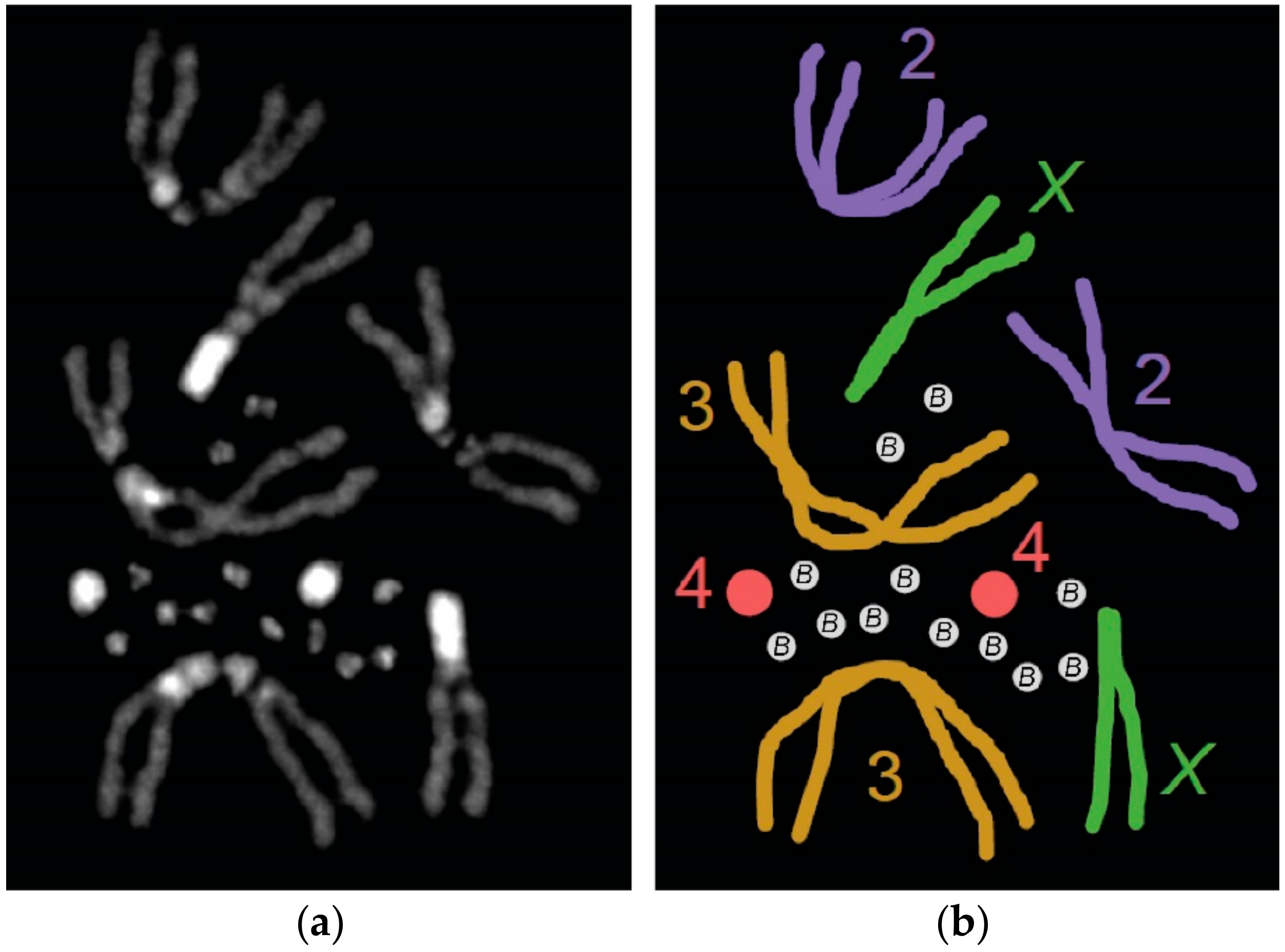
3.1. Characterization of the Original Drosophila melanogaster B chromosome
3.2. Effects of the Drosophila melanogaster B Chromosomes on the A Chromosomes
4. The D. melanogaster B Chromosome as an Emerging Model for B Chromosome Biology
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wilson, E.B. The supernumerary chromosomes of Hemiptera. Science 1907, 26, 870–871. [Google Scholar] [CrossRef]
- Jones, R.N.; Rees, H. B Chromosomes; Academic Press: London, UK, 1982; ISBN 9780123900609. [Google Scholar]
- Camacho, J. B chromosomes. In The Evolution of the Genome; Gregory, T.R., Ed.; Elsevier: San Diego, CA, USA, 2005; pp. 223–286. ISBN 9780123014634. [Google Scholar]
- D’Ambrosio, U.; Alonso-Lifante, M.P.; Barros, K.; Kovařík, A.; Mas de Xaxars, G.; Garcia, S. B-chrom: A database on B chromosomes of plants, animals and fungi. New Phytol. 2017, 216, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Banaei-Moghaddam, A.M.; Martis, M.M.; Macas, J.; Gundlach, H.; Himmelbach, A.; Altschmied, L.; Mayer, K.F.X.; Houben, A. Genes on B chromosomes: old questions revisited with new tools. Biochim. Biophys. Acta 2015, 1849, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Ruban, A.; Schmutzer, T.; Scholz, U.; Houben, A. How next-generation sequencing has aided our understanding of the sequence composition and origin of B chromosomes. Genes (Basel) 2017, 8, 294. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.T.; Nakajima, R.T.; Fantinatti, B.E.A.; Marques, D.F.; Almeida, R.O.; Simões, R.P.; Martins, C. B chromosomes: from cytogenetics to systems biology. Chromosoma 2017, 126, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Domínguez, B.; Ruiz-Ruano, F.J.; Cabrero, J.; Corral, J.M.; López-León, M.D.; Sharbel, T.F.; Camacho, J.P.M. Protein-coding genes in B chromosomes of the grasshopper Eyprepocnemis plorans. Sci. Rep. 2017, 7, 45200. [Google Scholar] [CrossRef] [PubMed]
- Coan, R.L.B.; Martins, C. Landscape of transposable elements focusing on the B chromosome of the cichlid fish Astatotilapia latifasciata. Genes (Basel) 2018, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Makunin, A.I.; Dementyeva, P.V.; Graphodatsky, A.S.; Volobouev, V.T.; Kukekova, A.V.; Trifonov, V.A. Genes on B chromosomes of vertebrates. Mol. Cytogenet. 2014, 7, 99. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N. Transmission and Drive Involving Parasitic B Chromosomes. Genes (Basel) 2018, 9, 388. [Google Scholar] [CrossRef] [PubMed]
- Bauerly, E.; Hughes, S.E.; Vietti, D.R.; Miller, D.E.; McDowell, W.; Hawley, R.S. Discovery of supernumerary B chromosomes in Drosophila melanogaster. Genetics 2014, 196, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, S.L.; Miller, D.E.; Eche, S.; Hawley, R.S. Origin, Composition, and Structure of the Supernumerary B Chromosome of Drosophila melanogaster. Genetics 2018. [Google Scholar] [CrossRef]
- O’Grady, P.M.; DeSalle, R. Phylogeny of the Genus Drosophila. Genetics 2018, 209, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Clayton, F.E. Published karyotypes of the Drosophilidae. Dros. Inf. Serv. 1998, 81, 5–125. [Google Scholar]
- Clyde, M. Chromosome IV variation in D. albomicans Duda. Dros. Inf. Serv. 1980, 55, 25–26. [Google Scholar]
- Hatsumi, M.; Kitagawa, O. Supernumerary chromosomes in Drosophila albomicans collected in Thailand. Abstr. Int. Congr. Entomol. 1980, 16, 124. [Google Scholar]
- Tonomura, Y.; Tobari, Y.N. Y-chromosome variation and B-chromosomes in Drosophila malerkotliana. Sci. Rep. Tokyo Women’s Christ. Univ. 1983, 32, 705–711. [Google Scholar]
- Sundaran, A.K.; Gupta, J.P. A new finding of B chromosomes in Drosophila kikkawai Burla. Rev. Bras. Genet. 1994, 17, 223–224. [Google Scholar]
- Gutknecht, J.; Sperlich, D.; Bachmann, L. A species specific satellite DNA family of Drosophila subsilvestris appearing predominantly in B chromosomes. Chromosoma 1995, 103, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Zeng, Q.; Qian, Y.; Li, C.; Yang, Y. Research on the karyotype and evolution of Drosophila melanogaster species group. J. Genet. Genom. 2007, 34, 196–213. [Google Scholar] [CrossRef]
- Mafla-Mantilla, A.B.; Romero, E.G. The heterochromatin of Drosophila inca, Drosophila yangana and Drosophila huancavilcae of the inca subgroup, repleta group. Dros. Inf. Serv. 2009, 92, 10–15. [Google Scholar]
- Ramachandra, N.B.; Ranganath, H.A. Supernumerary chromosomes in Drosophila nasuta albomicana. Experientia 1985, 41, 680–681. [Google Scholar] [CrossRef] [PubMed]
- Ramachandra, N.B.; Ranganath, H.A. Further studies on B-chromosomes in D. nasuta albomicana. Dros. Inf. Serv. 1985, 61, 139–140. [Google Scholar]
- Ramachandra, N.B.; Ranganath, H.A. Accumulation of B-chromosomes in Drosophila nasuta albomicana. Curr. Sci. 1987, 56, 850–852. [Google Scholar]
- Hatsumi, M. Karyotype polymorphism in Drosophila albomicans. Genome 1987, 29, 395–400. [Google Scholar] [CrossRef]
- Ramachandra, N.B.; Ranganath, H.A. Characterization of heterochromatin in the B chromosomes of Drosophila nasuta albomicana. Chromosoma 1987, 95, 223–226. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhu, H.; Huang, Q.; Zhao, L.; Zhang, G.; Roy, S.W.; Vicoso, B.; Xuan, Z.; Ruan, J.; Zhang, Y.; et al. Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans. BMC Genom. 2012, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Sundaran, A.K.; Gupta, J.P. Accumulation of B chromosomes in Drosophila kikkawai Burla. Cytobios 1994, 80, 211–215. [Google Scholar] [PubMed]
- Baimai, V.; Chumchong, C. Karyotype variation and geographic distribution of the three sibling species of the Drosophila kikkawai complex. Genetica 1980, 54, 113–120. [Google Scholar] [CrossRef]
- Brncic, D. Chromosomal structure of populations of Drosophila flavopilosa studied in larvae collected in their natural breeding sites. Chromosoma 1962, 13, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Bonner, A.M.; Hughes, S.E.; Chisholm, J.A.; Smith, S.K.; Slaughter, B.D.; Unruh, J.R.; Collins, K.A.; Friederichs, J.M.; Florens, L.; Swanson, S.K.; et al. Binding of Drosophila Polo kinase to its regulator Matrimony is noncanonical and involves two separate functional domains. Proc. Natl. Acad. Sci. USA 2013, 110, e1222–e1231. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.; Orme, C.; Kramer, J.; Namba, L.; Champion, M.; Palladino, M.J.; Natzle, J.; Hawley, R.S. A deficiency screen of the major autosomes identifies a gene (matrimony) that is haplo-insufficient for achiasmate segregation in Drosophila oocytes. Genetics 2003, 165, 637–652. [Google Scholar] [PubMed]
- Xiang, Y.; Takeo, S.; Florens, L.; Hughes, S.E.; Huo, L.-J.; Gilliland, W.D.; Swanson, S.K.; Teeter, K.; Schwartz, J.W.; Washburn, M.P.; et al. The inhibition of polo kinase by matrimony maintains G2 arrest in the meiotic cell cycle. PLoS Biol. 2007, 5, e323. [Google Scholar] [CrossRef] [PubMed]
- Dernburg, A.F.; Sedat, J.W.; Hawley, R.S. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 1996, 86, 135–146. [Google Scholar] [CrossRef]
- Karpen, G.H.; Le, M.H.; Le, H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science 1996, 273, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Hawley, R.S.; Theurkauf, W.E. Requiem for distributive segregation: A chiasmate segregation in Drosophila females. Trends Genet. 1993, 9, 310–317. [Google Scholar] [CrossRef]
- Elgin, S.C.R.; Reuter, G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. CSH Perspect. Biol. 2013, 5, a017780. [Google Scholar] [CrossRef] [PubMed]
- Page, A.W.; Orr-Weaver, T.L. Activation of the meiotic divisions in Drosophila oocytes. Dev. Biol. 1997, 183, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Endow, S.A.; Komma, D.J. Spindle dynamics during meiosis in Drosophila oocytes. J. Cell Biol. 1997, 137, 1321–1336. [Google Scholar] [CrossRef] [PubMed]
- Fuster, C.; Rigola, M.A.; Egozcue, J. Human supernumeraries: Are they B chromosomes? Cytogenet. Genome Res. 2004, 106, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T.; Mrasek, K.; Kosyakova, N.; Ogilvie, C.M.; Vermeesch, J.; Trifonov, V.; Rubtsov, N. Small supernumerary marker chromosomes (sSMC) in humans; are there B chromosomes hidden among them. Mol. Cytogenet. 2008, 1, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liehr, T. Small Supernumerary Marker Chromosomes (SMCs), A Guide for Human Geneticists and Clinicians; Springer: Berlin/Heidelberg, Germany, 2012; ISBN 9783642207655. [Google Scholar]

| Year Reported | Drosophila Species | Location of Sample Collection | Reference |
|---|---|---|---|
| 1980 | D. albomicans1 | Chiang Mai, Thailand | [16,17] |
| 1983 | D. malerkotliana | Multiple locations in Southeast Asia | [18] |
| 1994 | D. kikkawai | Bhubaneswar, India | [19] |
| 1995 | D. subsilvestris | Tübingen, Germany | [20] |
| 2007 | D. lini | Malipo county, Yunnan province, China | [21] |
| 2007 | D. pseudoananassae | Jianfengling, Hainan province, China | [21] |
| 2009 | D. yangana2 | Loja province, Ecuador | [22] |
| 2009 | D. huancavilcae2 | Manabí province, Ecuador | [22] |
| 2014 | D. melanogaster | Domesticated laboratory stock | [12] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanlon, S.L.; Hawley, R.S. B Chromosomes in the Drosophila Genus. Genes 2018, 9, 470. https://doi.org/10.3390/genes9100470
Hanlon SL, Hawley RS. B Chromosomes in the Drosophila Genus. Genes. 2018; 9(10):470. https://doi.org/10.3390/genes9100470
Chicago/Turabian StyleHanlon, Stacey L., and R. Scott Hawley. 2018. "B Chromosomes in the Drosophila Genus" Genes 9, no. 10: 470. https://doi.org/10.3390/genes9100470
APA StyleHanlon, S. L., & Hawley, R. S. (2018). B Chromosomes in the Drosophila Genus. Genes, 9(10), 470. https://doi.org/10.3390/genes9100470





