The E3 Ubiquitin Ligase TRIM21 Promotes HBV DNA Polymerase Degradation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Transfections
2.2. FLAG Affinity Purification
2.3. Coimmunoprecipitation and Glutathione S transferase (GST) pull-down assay
2.4. Western Blot
2.5. Immunofluorescence
2.6. Ubiquitination Assay
2.7. HBV DNA Analysis
2.8. Southern Blot Assay
2.9. Statistical Analysis
2.10. Plasmid Construction
2.11. RNA Preparation and Reverse Transcription Quantitative PCR (RT-qPCR)
3. Results
3.1. TRIM21 Interacts with HBV DNA Pol
3.2. TRIM21 Interacts with the TP Domain of HBV DNA Pol via its SPRY Domain
3.3. TRIM21 Negatively Regulates the Stability of HBV DNA Pol
3.4. TRIM21 Promotes the Degradation of HBV DNA Pol through the Ubiquitin Proteasome Pathway via Its RING Domain
3.5. TRIM21 Mediates the K48-Linked Ubiquitination of HBV DNA Pol at K260 and K283
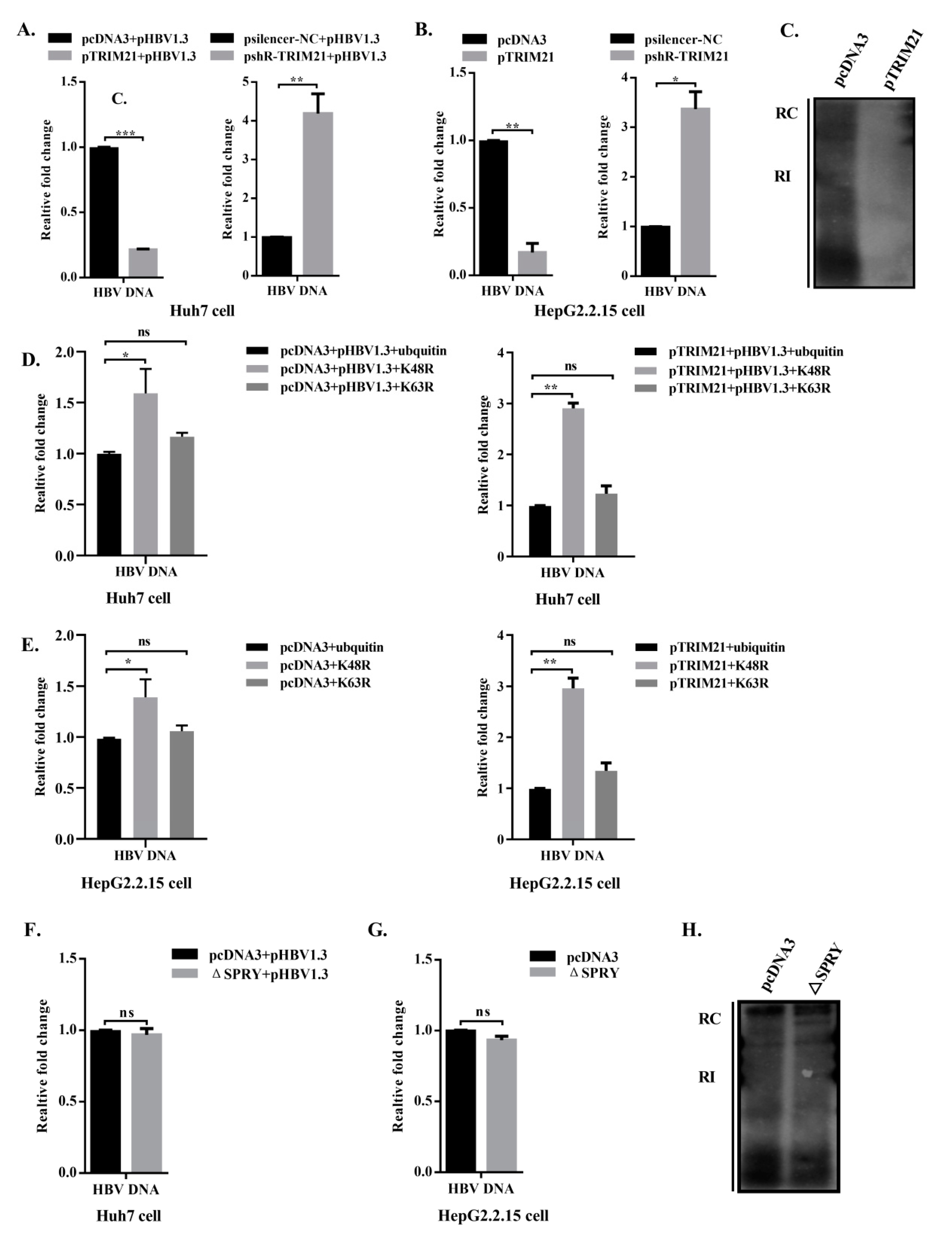
3.6. TRIM21 Restricts HBV DNA Replication, and the SPRY Domain is Crucial for the Antiviral Role of TRIM21
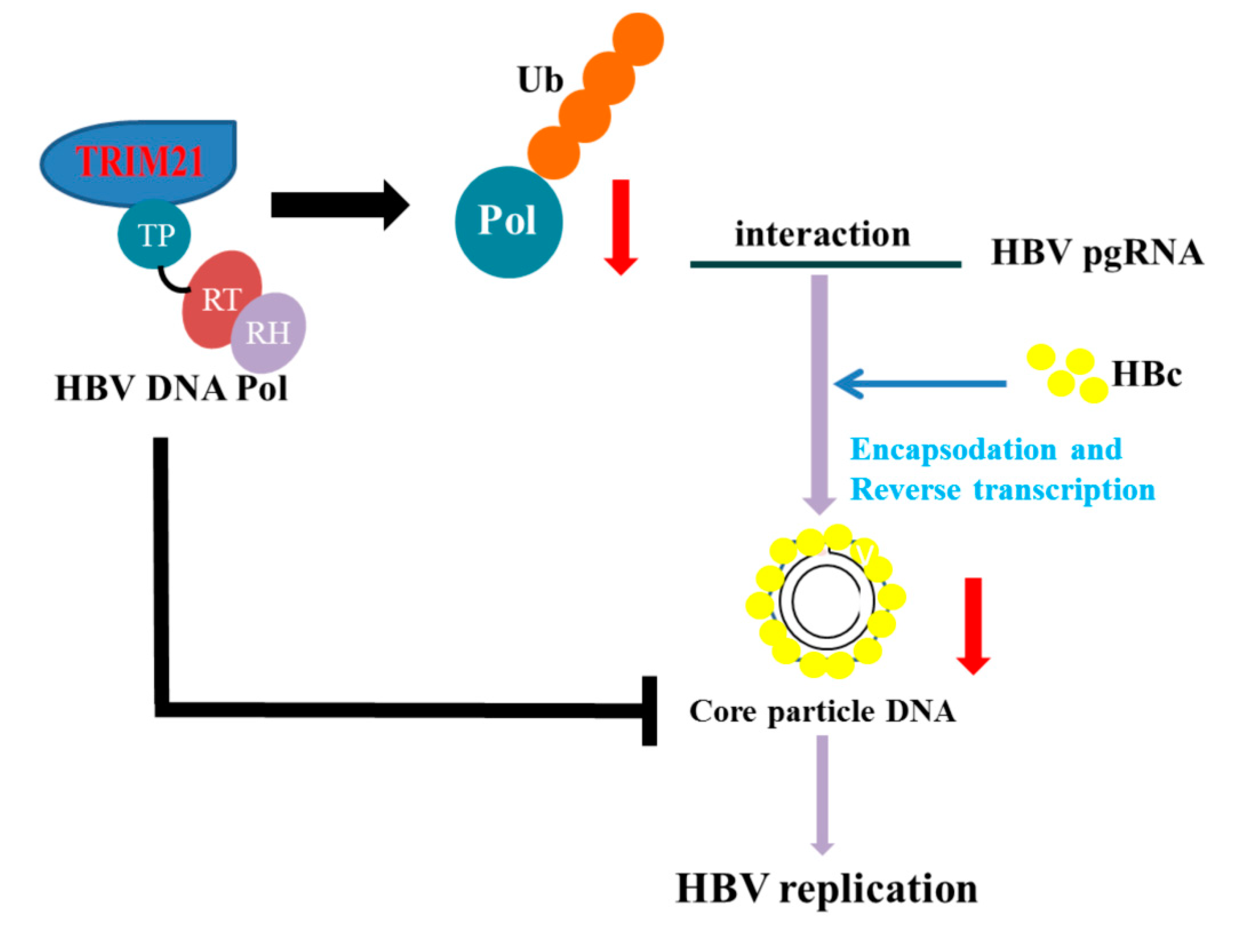
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Ortega-Prieto, A.M.; Skelton, J.K.; Wai, S.N.; Large, E.; Lussignol, M.; Vizcay-Barrena, G.; Hughes, D.; Fleck, R.A.; Thursz, M.; Catanese, M.T.; et al. 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection. Nat. Commun. 2018, 9, 682. [Google Scholar] [CrossRef] [Green Version]
- Michailidis, E.; Pabon, J.; Xiang, K.; Park, P.; Ramanan, V.; Hoffmann, H.H.; Schneider, W.M.; Bhatia, S.N.; Jong, Y.P.; Shlomai, A. A robust cell culture system supporting the complete life cycle of hepatitis B virus. Sci. Rep. 2017, 7, 16616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burwitz, B.J.; Wettengel, J.M.; Mückhäusl, M.A.; Ringelhan, M.; Ko, C.; Festag, M.M.; Hammond, K.B.; Northrup, M.; Bimber, B.N.; Jacob, T. Hepatocytic expression of human sodium-taurocholate cotransporting polypeptide enables hepatitis B virus infection of macaques. Nat. Commun. 2017, 8, 2146. [Google Scholar] [CrossRef] [PubMed]
- Schlicksup, C.J.; Wang, C.Y.; Francis, S.; Venkatakrishnan, B.; Turner, W.W.; Vannieuwenhze, M.; Zlotnick, A. Hepatitis B virus core protein allosteric modulators can distort and disrupt intact capsids. eLife Sci. 2018, 7, e31473. [Google Scholar] [CrossRef] [PubMed]
- Rajbhandari, R.; Chung, R.T. Treatment of Hepatitis B: A Concise Review. Clin. Transl. Gastroenterol. 2016, 7, e190. [Google Scholar] [CrossRef]
- Lucifora, J.; Xia, Y.; Reisinger, F.; Zhang, K.; Stadler, D.; Cheng, X.; Sprinzl, M.F.; Koppensteiner, H.; Makowska, Z.; Volz, T. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014, 343, 1221–1228. [Google Scholar] [CrossRef]
- Lamontagne, R.J.; Bagga, S.; Bouchard, M.J. Hepatitis B virus molecular biology and pathogenesis. Hepatoma Res. 2016, 2, 163–186. [Google Scholar] [CrossRef]
- Schädler, S.; Hildt, E. HBV Life Cycle: Entry and Morphogenesis. Viruses 2009, 1, 185–209. [Google Scholar] [CrossRef]
- Yang, X.; Li, H.; Sun, H.; Fan, H.; Hu, Y.; Liu, M.; Li, X.; Tang, H. Hepatitis B virus-encoded miRNA controls viral replication. J. Virol. 2017, 91, e01919-16. [Google Scholar] [CrossRef] [Green Version]
- Clark, D.N.; Flanagan, J.M.; Hu, J. Mapping of Functional Subdomains in the Terminal Protein Domain of Hepatitis B Virus Polymerase. J. Virol. 2017, 91, e01785-16. [Google Scholar] [CrossRef] [Green Version]
- Nassal, M. HBV cccDNA: Viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 2015, 64, 1972–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S.A.; Hu, J. Protein-Primed Terminal Transferase Activity of Hepatitis B Virus Polymerase. J. Virol. 2013, 87, 2563–2576. [Google Scholar]
- Chung, H.J.; Chen, X.; Yu, Y.; Lee, H.K.; Song, C.H.; Choe, H.; Lee, S.; Kim, H.J.; Hong, S.T. A critical role of hepatitis B virus polymerase in cirrhosis, hepatocellular carcinoma, and steatosis. Febs. Open Biol. 2018, 8, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.N.; Hu, J. Unveiling the roles of HBV polymerase for new antiviral strategies. Future Virol. 2015, 10, 283–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senft, D.; Qi, J.; Ronai, Z.E.A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nature Reviews Cancer 2017, 18, 69–88. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; David, K. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, A.L.; Ciechanover, A. Targeting Proteins for Destruction by the Ubiquitin System: Implications for Human Pathobiology. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 73–96. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Chai, Q.Y.; Liu, C.H. The ubiquitin system: A critical regulator of innate immunity and pathogen|[ndash]|host interactions. Cell. Mol. Immunol. 2016, 13, 560–576. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Sun, S.C. Ubiquitin signaling in immune responses. Cell Res. 2016, 26, 457–483. [Google Scholar] [CrossRef] [Green Version]
- Rajsbaum, R.; Garcíasastre, A.; Versteeg, G.A. TRIMmunity: The roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J. Mol. Biol. 2014, 426, 1265–1284. [Google Scholar] [CrossRef] [Green Version]
- Sparrer, K.M.J.; Gack, M.U. TRIM proteins: New players in virus-induced autophagy. PLoS Pathog 2018, 14, e1006787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wu, H.; Wu, W.; Zhuo, W.; Liu, W.; Zhang, Y.; Cheng, M.; Chen, Y.G.; Gao, N.; Yu, H. Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res. 2014, 24, 762–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Chen, Y.; Li, C.; Wu, Y.; Guo, L.; Peng, C.; Huang, Y.; Cheng, G.; Qin, F.X. TRIM14 inhibits hepatitis C virus infection by SPRY domain-dependent targeted degradation of the viral NS5A protein. Sci. Rep. 2016, 6, 32336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Zhao, X.; Sun, D.; Yang, L.; Chong, C.; Pan, Y.; Chi, X.; Gao, Y.; Wang, M.; Shi, X.; et al. Interferon alpha (IFNalpha)-induced TRIM22 interrupts HCV replication by ubiquitinating NS5A. Cell Mol. Immunol. 2016, 13, 94–102. [Google Scholar] [CrossRef] [Green Version]
- Gack, M.U.; Shin, Y.C.; Joo, C.H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef]
- Fan, W.; Wu, M.; Qian, S.; Yun, Z.; Chen, H.; Li, X.; Ping, Q. TRIM52 inhibits Japanese Encephalitis Virus replication by degrading the viral NS2A. Sci. Rep. 2016, 6, 33698. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Zhang, M.; Chu, H.; Zhang, H.; Wu, H.; Song, G.; Wang, P.; Zhao, K.; Hou, J.; Wang, X. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat. Immunol. 2016, 18, 214–224. [Google Scholar] [CrossRef]
- Seo, G.J.; Kim, C.; Shin, W.J.; Sklan, E.H.; Eoh, H.; Jung, J.U. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat. Commun. 2018, 9, 613. [Google Scholar] [CrossRef] [Green Version]
- Tan, G.; Xu, F.; Song, H.; Yuan, Y.; Xiao, Q.; Ma, F.; Qin, F.X.-F.; Cheng, G. Identification of TRIM14 as a Type I IFN-Stimulated Gene Controlling Hepatitis B Virus Replication by Targeting HBx. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Gao, B.; Duan, Z.W. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 2010, 50, 424–433. [Google Scholar] [CrossRef]
- Tan, G.; Xiao, Q.; Song, H.; Ma, F.; Xu, F.; Peng, D.; Li, N.; Wang, X.; Niu, J.; Gao, P. Type I IFN augments IL-27-dependent TRIM25 expression to inhibit HBV replication. Cell. Mol. Immunol. 2017, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.S.; Yang, M.C.W.; Wang, B.; Weissler, J.C. Autoantigen Ro52 directly interacts with human IgG heavy chain in vivo in mammalian cells. Mol. Immunol. 2000, 37, 591–602. [Google Scholar] [CrossRef]
- Mcewan, W.A.; Tam, J.C.; Watkinson, R.E.; Bidgood, S.R.; Mallery, D.L.; James, L.C. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat. Immunol. 2013, 14, 327–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Bao, M.; Lu, N.; Weng, L.; Yuan, B.; Liu, Y.J. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol. 2013, 14, 172–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.; Dinh, P.X.; Pattnaik, A.K. Trim21 regulates Nmi-IFI35 complex-mediated inhibition of innate antiviral response. Virology 2015, 485, 383–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watkinson, R.E.; Mcewan, W.A.; Tam, J.C.H.; Vaysburd, M.; James, L.C. TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized Virus. PLoS Pathog. 2015, 11, e1005253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seeger, C.; Mason, W.S. Molecular biology of hepatitis B virus infection. Virology 2015, 479–480, 672–686. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.J.; Sun, L.J. Nonproteolytic Functions of Ubiquitin in Cell Signaling. Mol. Cell 2009, 33, 275–286. [Google Scholar] [CrossRef]
- Kovalskyy, D.B.; Ivanov, D.N. Recognition of the HIV Capsid by the TRIM5α Restriction Factor Is Mediated by a Subset of Pre-Existing Conformations of the TRIM5α SPRY Domain. Biochemistry 2014, 53, 1466–1476. [Google Scholar] [CrossRef] [Green Version]
- She, S.; Yang, M.; Hu, H.; Hu, P.; Yang, Y.; Ren, H. Proteomics Based Identification of Autotaxin As An Anti-Hepatitis B Virus Factor and a Promoter of Hepatoma Cell Invasion and Migration. Cell. Physiol. Biochem. 2018, 45, 744–760. [Google Scholar] [CrossRef]
- Iloeje, U.H.; Yang, H.I.; Su, J.; Jen, C.L.; You, S.L.; Chen, C.J. Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology 2006, 130, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Yang, H.I.; Liu, J.; Batrlautermann, R.; Jen, C.L.; Iloeje, U.H.; Lu, S.N.; You, S.L.; Wang, L.Y.; Chen, C.J. Prediction models of long-term cirrhosis and hepatocellular carcinoma risk in chronic hepatitis B patients: Risk scores integrating host and virus profiles. Hepatology 2013, 58, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Gish, R.G.; Given, B.D.; Lai, C.L.; Locarnini, S.A.; Lau, J.Y.; Lewis, D.L.; Schluep, T. Chronic hepatitis B: Virology, natural history, current management and a glimpse at future opportunities. Antivir. Res. 2015, 121, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.A.; Hu, J. Hepatitis B virus reverse transcriptase: Diverse functions as classical and emerging targets for antiviral intervention. Emerg. Microbes Infect. 2013, 2, e56. [Google Scholar] [CrossRef]
- Park, S.G.; Jung, G. Human hepatitis B virus polymerase interacts with the molecular chaperonin Hsp60. J. Virol. 2001, 75, 6962–6968. [Google Scholar] [CrossRef] [Green Version]
- Cho, G.; Park, S.G.; Jung, G. Localization of HSP90 binding sites in the human hepatitis B virus polymerase. Biochem. Biophys. Res. Commun. 2000, 269, 191–196. [Google Scholar] [CrossRef]
- Sung, G.P.; Jin, K.R.; Guhung, J. Hsp90 makes the human HBV Pol competent for in vitro priming rather than maintaining the human HBV Pol/pregenomic RNA complex. Arch. Biochem. Biophys. 2002, 401, 99–107. [Google Scholar]
- Buetow, L.; Huang, D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nature Rev. Mol. Cell Biol. 2016, 17, 626–642. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xie, P.; Lu, L.; Wang, J.; Diao, L.; Liu, Z.; Guo, F.; He, Y.; Liu, Y.; Huang, Q. An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network. Nat. Commun. 2017, 8, 347. [Google Scholar] [CrossRef] [Green Version]
- Grossegesse, M.; Doellinger, J.; Fritsch, A.; Laue, M.; Piesker, J.; Schaade, L.; Nitsche, A. Global ubiquitination analysis reveals extensive modification and proteasomal degradation of cowpox virus proteins, but preservation of viral cores. Sci. Rep. 2018, 8, 1807. [Google Scholar] [CrossRef]
- Giraldo, M.I.; Vargas-Cuartas, O.; Gallego-Gomez, J.C.; Shi, P.Y.; Padilla-Sanabria, L.; Castaño-Osorio, J.C.; Rajsbaum, R. K48-linked polyubiquitination of dengue virus NS1 protein inhibits its interaction with the viral partner NS4B. Virus Res. 2018, 246, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Jin, F.; Chang, L.; Yang, Y.; Peng, H.; Duan, C. NIRF, a novel ubiquitin ligase, interacts with hepatitis B virus core protein and promotes its degradation. Biotechnol. Lett. 2012, 34, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ni, J.; Li, J.; Shi, B.; Xu, Y.; Yuan, Z. Inhibition of hepatitis B virus replication by cIAP2 involves accelerating the ubiquitin-proteasome-mediated destruction of polymerase. J. Virol. 2011, 85, 11457–11467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, L.; Zhao, J.; Gao, S.; Ji, T.; Song, T.; Li, Y.; Wang, J.; Geng, C.; Long, M.; Chen, J.; et al. Restriction of hepatitis B virus replication by c-Abl–induced proteasomal degradation of the viral polymerase. Sci. Adv. 2019, 5, eaau7130. [Google Scholar] [CrossRef] [Green Version]
- Esposito, D.; Koliopoulos, M.G.; Rittinger, K. Structural determinants of TRIM protein function. Biochem. Soc. Trans. 2017, 45, 183–191. [Google Scholar] [CrossRef]
- Short, K.M.; Cox, T.C. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem. 2006, 281, 8970–8980. [Google Scholar] [CrossRef] [Green Version]
- James, L.C.; Keeble, A.H.; Khan, Z.; Rhodes, D.A.; Trowsdale, J. Structural Basis for PRYSPRY-Mediated Tripartite Motif (TRIM) Protein Function. Proc. Natl. Acad. Sci. USA 2007, 104, 6200–6205. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Wang, J.; Sun, B. Trim21: A novel negative regulator in DNA sensor signaling. Cell. Mol. Immunol. 2013, 10, 190–192. [Google Scholar] [CrossRef] [Green Version]
- Unchwaniwala, N.; Sherer, N.M.; Loeb, D.D. Hepatitis B virus polymerase localizes to the mitochondria and its terminal protein domain contains the mitochondrial-targeting signal. J. Virol. 2016, 90, 8705–8719. [Google Scholar] [CrossRef] [Green Version]
- Chunkyu, K.; Youn-Chul, S.; Woo-Jin, P.; Seungtaek, K.; Jonghwa, K.; Wang-Shick, R. Residues Arg703, Asp777, and Arg781 of the RNase H domain of hepatitis B virus polymerase are critical for viral DNA synthesis. J. Virol. 2014, 88, 154–163. [Google Scholar]
- Tu, T.; Budzinska, M.A.; Shackel, N.A.; Urban, S. HBV DNA Integration: Molecular Mechanisms and Clinical Implications. Viruses 2017, 9, E75. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.; Nassal, M. Hepatitis B virus replication. World J. Gastroenterol. 2007, 3, 217–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, Y.C.; Park, S.; Ryu, W.S. A conserved arginine residue in the terminal protein domain of hepatitis B virus polymerase is critical for RNA pre-genome encapsidation. J. Gen. Virol. 2011, 92, 1809–1816. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Chen, J.; Li, Y.; Wang, W.; Du, X.; Song, W.; Zhang, W.; Lin, L.; Yuan, Z. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J. Virol. 2015, 89, 2287–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sjöstrand, M.; Ambrosi, A.; Brauner, S.; Sullivan, J.; Malin, S.; Kuchroo, V.K.; Espinosa, A.; Wahrenherlenius, M. Expression of the immune regulator tripartite-motif 21 is controlled by IFN regulatory factors. J. Immunol. 2013, 191, 3753–3763. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Usage | Primer Name | Sequence (5′–3′) |
|---|---|---|
| Construction for HBV DNA Pol | HBV-DNA- Pol-F | GCCGGTACCGCCATGCCCCTATCTTATCCACAC |
| HBV-DNA- Pol-R | ATAAGAATGCGGCCGCTCACGGGGGTCTCCATGCGAC | |
| Construction for HBV DNA Pol-mut | HBV-DNA-Pol-K260R-F | CCAGCAGATCCGCCTCCTGCCTCCACCAAT |
| HBV-DNA-Pol-K260R-R | GCAGGAGGCGGATCTGCTGGCAAAGTTTGT | |
| HBV-DNA-Pol-K283R-F | TGAGAGACACTCATCCTCAGGCCATGCAGT | |
| HBV-DNA-Pol-K283R-R | CTGAGGATGAGTGTCTCTCAAAGGTGGAGA | |
| Construction for HBV DNA Pol- del-mut | HBV-P-del- TP-mut-F | GCCGGTACCATGTCTACAGCATGGGGCAGAATC |
| HBV-P-del- RT-mut-F | CTACTGCCTCTCCCTTATCGTCAATCTTCTCGAGCCAGGTCTGTGCCAAGTGTTTG | |
| HBV-P-del- RT-mut-R | CTGGCTCGAGAAGATTGACGATAAGGGAGAGGCAGTAG | |
| HBV-P-del- RH-mut-R | ATAAGAATGCGGCCGCTTACCGTTGCCGGGCAACGGGGTA | |
| Construction for pHBV1.3 | pHBV1.3 | GGGTTTCGCCACCTCTGACT |
| Construction for TRIM21 | TRIM21-F | GCGGAATTCATGGCTTCAGCAGCACGCTTG |
| TRIM21-R | GCTCACCTCGAGTAGTCAGTGGATCCTTGTGATC | |
| Construction for shR-TRIM21 | shR-TRIM21-F | GATCCGCAGCACGCTTGACAATGATGCTCGAGCATCATTGTCAAGCGTGCTGCTTTTTGA |
| shR-TRIM21-R | AGCTTCAAAAAGCAGCACGCTTGACAATGATGCTCGAGCATCATTGTCAAGCGTGCTGCG | |
| Construction for TRIM21 -del-mut | TRIM21-del-RING-F | GCGGAATTCATGCTGCTCAAGAATCTCCGGCCC |
| TRIM21-del-CC-F | ACGCCATGGTCCCTCTTGAGGAGGAGCTCATCTCAGAGCTAGATCGAAGGTGC | |
| TRIM21-del-CC-R | GCACCTTCGATCTAGCTCTGAGATGAGCTCCTCCTCAAGAGGGACCATGGCGT | |
| TRIM21-del-PRY-F | AAGATGCTGAGGACATGTGCACAGCACTTTCACTCTGGAAAACATTACTGG | |
| TRIM21-del-PRY-R | CCAGTAATGTTTTCCAGAGTGAAAGTGCTGTGCACATGTCCTCAGCATCTT | |
| TRIM21-del-SPRY-R | GCTCACCTCGAGTCACTGGGCACCCAGGACCATAGG | |
| TRIM21-del-PRY-SPRY-R | GCTCACCTCGAGTCACTGTAGGGCCTGGCTCTGCTG | |
| Construction for Ubiquitin | Ubiquitin-F | GACGAATTCATGCAGATCTTCGTGAAAAC |
| Ubiquitin-R | GCTCACCTCGAGCTAACCACCTCTCAGACGCAGGAC | |
| Construction for Ubiquitin -mut | Ubiquitin- K48R-F | CTCATCTTTGCAGGCAGGCAGCTGGAAGAT |
| Ubiquitin- K48R-R | ATCTTCCAGCTGCCTGCCTGCAAAGATGAG | |
| Ubiquitin- K63R-R | GCACCTCGAGCTAACCACCTCTCAGACGCAGGACCAGGTGCAGGGTCGACTCCCTCTGGA | |
| RT-qPCR for TRIM21 | TRIM21-qRT-F | CAGTCCTGGTTTCAATGATG |
| TRIM21-qRT-R | TGGGCCAAATATAAGGAGGC | |
| RT-qPCR for HBV DNA Pol | Pol-qRT-F | CTCTCTTTACGCGGACTCCC |
| Pol-qRT-R | TCTTCTAGGGGACCTGCCTC | |
| qPCR for HBV DNA copies | HBV DNA copies -F | TCTTGCCTTACTTTTGGAAG |
| HBV DNA copies -R | AGTTCTTCTTCTAGGGGACC | |
| RT-qPCR for β-actin | β-actin-F | CGTGACATTAAGGAGAAGCTG |
| β-actin-R | CTAGAAGCATTTGCGGTGGAC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, T.; Zhao, X.; Zhu, Y.; Fan, H.; Tang, H. The E3 Ubiquitin Ligase TRIM21 Promotes HBV DNA Polymerase Degradation. Viruses 2020, 12, 346. https://doi.org/10.3390/v12030346
Mu T, Zhao X, Zhu Y, Fan H, Tang H. The E3 Ubiquitin Ligase TRIM21 Promotes HBV DNA Polymerase Degradation. Viruses. 2020; 12(3):346. https://doi.org/10.3390/v12030346
Chicago/Turabian StyleMu, Ting, Xiaoqing Zhao, Yanan Zhu, Hongxia Fan, and Hua Tang. 2020. "The E3 Ubiquitin Ligase TRIM21 Promotes HBV DNA Polymerase Degradation" Viruses 12, no. 3: 346. https://doi.org/10.3390/v12030346
APA StyleMu, T., Zhao, X., Zhu, Y., Fan, H., & Tang, H. (2020). The E3 Ubiquitin Ligase TRIM21 Promotes HBV DNA Polymerase Degradation. Viruses, 12(3), 346. https://doi.org/10.3390/v12030346




