PGC-1α in Disease: Recent Renal Insights into a Versatile Metabolic Regulator
Abstract
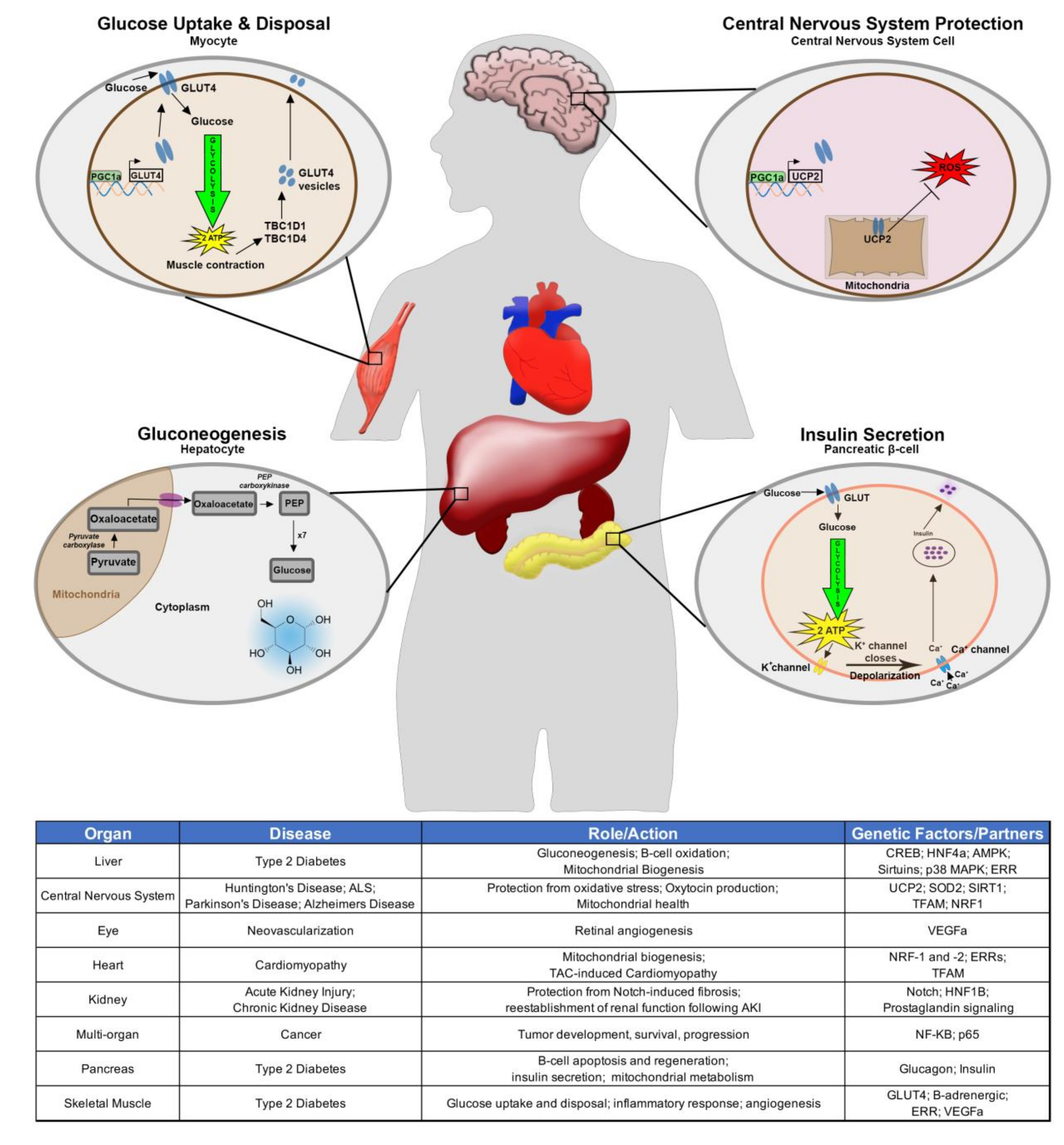
1. Introduction
2. PGC-1α in the Context of Human Disease and Development
3. PGC-1α in Kidney Disease and Development
3.1. Kidney Cancer
3.2. Acute Kidney Injury (AKI)
3.3. Chronic Kidney Disease (CKD)
3.4. Future Direction: Polycystic Kidney Disease (PKD) and Cilia
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Puigserver, P.; Wu, Z.; Park, C.W.; Graves, R.; Wright, M.; Spiegelman, B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998, 92, 829–839. [Google Scholar] [CrossRef]
- Knutti, D.; Kaul, A.; Kralli, A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol. Cell. Biol. 2000, 20, 2411–2422. [Google Scholar] [CrossRef]
- Lin, J.; Handschin, C.; Spiegelman, B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005, 1, 361–370. [Google Scholar] [CrossRef]
- Handschin, C.; Spiegelman, B.M. Peroxisome proliferator-activated receptor γ coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006, 27, 728–735. [Google Scholar] [CrossRef]
- Fernandez-Marcos, P.J.; Auwerx, J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 2011, 93, 884S–890S. [Google Scholar] [CrossRef]
- Patten, I.S.; Arany, Z. PGC-1 coactivators in the cardiovascular system. Trends Endocrinol. Metab. 2012, 23, 90–97. [Google Scholar] [CrossRef]
- Liu, C.; Lu, Y. PGC-1 coactivators in the control of energy metabolism. Acta Biochim. Biophys. Sin. 2011, 43, 248–257. [Google Scholar] [CrossRef]
- Lin, J.; Puigserver, P.; Donovan, J.; Tarr, P.; Spiegelman, B.M. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2001, 277, 1645–1648. [Google Scholar] [CrossRef]
- Andersson, U.; Scarpulla, R.C. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell. Biol. 2001, 21, 3738–3749. [Google Scholar] [CrossRef]
- Tiefenböck, S.K.; Baltzer, C.; Egli, N.A.; Frei, C. The Drosophila PGC-1 homologue Spargel coordinates mitochondrial activity to insulin signalling. EMBO J. 2009, 29, 171–183. [Google Scholar] [CrossRef]
- Li, S.; Liu, C.; Li, N.; Hao, T.; Han, T.; Hill, D.E.; Vidal, M.; Lu, Y. Genome-wide coactivation analysis of PGC-1α identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008, 8, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wu, P.-H.; Tarr, P.T.; Lindenberg, K.S.; St-Pierre, J.; Zhang, C.-Y.; Mootha, V.K.; Jager, S.; Vianna, C.R.; Reznick, R.M.; et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell 2004, 119, 121–135. [Google Scholar] [CrossRef]
- Boss, O.; Bachman, E.; Vidal-Puig, A.; Zhang, C.-Y.; Peroni, O.; Lowell, B.B. Role of the β3-adrenergic receptor and/or a putative β4-adrenergic receptor on the expression of uncoupling proteins and peroxisome proliferator-activated receptor-γ coactivator-1. Biochem. Biophys. Res. Commun. 1999, 261, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ambrosi, J.; Frühbeck, G.; Martínez, J.A. Rapid in vivo PGC-1 mRNA upregulation in brown adipose tissue of Wistar rats by a β3-adrenergic agonist and lack of effect of leptin. Mol. Cell. Endocrinol. 2001, 176, 85–90. [Google Scholar] [CrossRef]
- Handschin, C.; Rhee, J.; Lin, J.; Tarr, P.T.; Spiegelman, B.M. An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc. Natl. Acad. Sci. USA 2003, 100, 7111–7116. [Google Scholar] [CrossRef]
- Cao, W.; Daniel, K.W.; Robidoux, J.; Puigserver, P.; Medvedev, A.; Bai, X.; Floering, L.M.; Spiegelman, B.M.; Collins, S. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 2004, 24, 3057–3067. [Google Scholar] [CrossRef]
- Yoon, J.C.; Puigserver, P.; Chen, G.; Donovan, J.; Wu, Z.; Rhee, J.; Adelmant, G.; Stafford, J.M.; Kahn, C.R.; Granner, D.K.; et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 2001, 413, 131–138. [Google Scholar] [CrossRef]
- Estall, J.L.; Kahn, M.; Cooper, M.P.; Fisher, F.M.; Wu, M.K.; Laznik, D.; Qu, L.; Cohen, D.E.; Shulman, G.I.; Spiegelman, B.M. Sensitivity of lipid metabolism and insulin signaling to genetic alterations in hepatic peroxisome proliferator-activated receptor-gamma coactivator-1alpha expression. Diabetes 2009, 58, 1499–1508. [Google Scholar] [CrossRef]
- Herzig, S.; Long, F.; Jhala, U.S.; Hedrick, S.; Quinn, R.; Bauer, A.; Rudolph, D.; Schutz, G.; Yoon, C.; Puigserver, P.; et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 2001, 413, 179–183. [Google Scholar] [CrossRef]
- Rhee, J.; Inoue, Y.; Yoon, J.C.; Puigserver, P.; Fan, M.; Gonzalez, F.J.; Spiegelman, B.M. Regulation of hepatic fasting response by PPAR coactivator-1 (PGC-1): Requirement for hepatocyte nuclear factor 4 in gluconeogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 4012–4017. [Google Scholar] [CrossRef]
- Puigserver, P.; Rhee, J.; Donovan, J.; Walkey, C.J.; Yoon, J.C.; Oriente, F.; Kitamura, Y.; Altomonte, J.; Dong, H.H.; Accili, M.; et al. Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1α interaction. Nature 2003, 423, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Leone, T.C.; Lehman, J.J.; Finck, B.N.; Schaeffer, P.J.; Wende, A.R.; Boudina, S.; Courtois, M.; Wozniak, D.F.; Sambandam, N.; Bernal-Mizrachi, C.; et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005, 3, e101. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Collins, Q.F.; Becker, T.C.; Robidoux, J.; Lupo, E.G.; Xiong, Y.; Daniel, K.W.; Floering, L.; Collins, S. p38 mitogen-activated protein kinase plays a stimulatory role in hepatic gluconeogenesis. J. Biol. Chem. 2005, 280, 42731–42737. [Google Scholar] [CrossRef] [PubMed]
- Besse-Patin, A.; Jeromson, S.; Levesque-Damphousse, P.; Secco, B.; Laplante, M.; Estall, J.L. PGC1A regulates the IRS1:IRS2 ratio during fasting to influence hepatic metabolism downstream of insulin. Proc. Natl. Acad. Sci. USA 2019, 116, 4285–4290. [Google Scholar] [CrossRef]
- Wu, H.; Kanatous, S.B.; Thurmond, F.A.; Gallardo, T.; Isotani, E.; Bassel-Duby, R.; Williams, R.S. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 2002, 296, 349–352. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, X.; Feng, Y.; Handschin, C.; Gullicksen, P.S.; Bare, O.; Labow, M.; Spiegelman, B.M.; Stevenson, S.C.; Feng, Y.; et al. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1 transcription and mitochondrial biogenesis in muscle cells. Proc. Natl. Acad. Sci. USA 2006, 103, 14379–14384. [Google Scholar] [CrossRef]
- Akimoto, T.; Sorg, B.S.; Yan, Z. Real-time imaging of peroxisome proliferator-activated receptor-γ coactivator-1α promoter activity in skeletal muscles of living mice. Am. J. Physiol. Physiol. 2004, 287, C790–C796. [Google Scholar] [CrossRef]
- Zhao, M.; New, L.; Kravchenko, V.V.; Kato, Y.; Gram, H.; Di Padova, F.; Olson, E.N.; Ulevitch, R.; Han, J. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 1999, 19, 21–30. [Google Scholar] [CrossRef]
- Akimoto, T.; Pohnert, S.C.; Li, P.; Zhang, M.; Gumbs, C.; Rosenberg, P.B.; Williams, R.S.; Yan, Z. Exercise stimulates PGC-1α transcription in skeletal muscle through activation of the p38 MAPK pathway. J. Biol. Chem. 2005, 280, 19587–19593. [Google Scholar] [CrossRef]
- Wu, H.; Deng, X.; Shi, Y.; Su, Y.; Wei, J.; Duan, H. PGC-1α, glucose metabolism and type 2 diabetes mellitus. J. Endocrinol. 2016, 229, R99–R115. [Google Scholar] [CrossRef]
- Yoon, J.; Xu, G.; Deeney, J.T.; Yang, S.-N.; Rhee, J.; Puigserver, P.; Levens, A.R.; Yang, R.; Zhang, C.-Y.; Lowell, B.B.; et al. Suppression of β cell energy metabolism and insulin release by PGC-1α. Dev. Cell 2003, 5, 73–83. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, M.; Deng, B.; Frontera, J.R.; Kover, K.L.; Aires, D.; Ding, H.; Carlson, S.E.; Turk, J.; Wang, W.; et al. Beta-cell injury in Ncb5or-null mice is exacerbated by consumption of a high-fat diet. Eur. J. Lipid Sci. Technol. 2011, 114, 233–243. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, J.-W.; Park, S.-Y.; You, Y.-H.; Ham, D.-S.; Park, H.-S.; Lee, S.-H.; Yang, H.K.; Yoon, K.-H. Targeting PGC-1α to overcome the harmful effects of glucocorticoids in porcine neonatal pancreas cell clusters. Transplantation 2014, 97, 273–279. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.T.; Gasparetti, A.L.; Pereira-Da-Silva, M.; Araújo, E.P.; Carvalheira, J.B.; Saad, M.J.A.; Boschero, A.C.; Carneiro, E.M.; Velloso, L.A. Peroxisome proliferator-activated receptor γ coactivator-1-dependent uncoupling protein-2 expression in pancreatic islets of rats: A novel pathway for neural control of insulin secretion. Diabetologia 2003, 46, 1522–1531. [Google Scholar] [CrossRef][Green Version]
- Li, N.; Brun, T.; Cnop, M.; Cunha, D.A.; Eizirik, D.L.; Maechler, P. Transient oxidative stress damages mitochondrial machinery inducing persistent β-cell dysfunction. J. Biol. Chem. 2009, 284, 23602–23612. [Google Scholar] [CrossRef]
- Wende, A.R.; Schaeffer, P.J.; Parker, G.J.; Zechner, C.; Han, D.-H.; Chen, M.M.; Hancock, C.R.; Lehman, J.J.; Huss, J.M.; McClain, D.A.; et al. A role for the transcriptional coactivator PGC-1α in muscle refueling. J. Biol. Chem. 2007, 282, 36642–36651. [Google Scholar] [CrossRef]
- Eisele, P.S.; Salatino, S.; Sobek, J.; Hottiger, M.O.; Handschin, C. The peroxisome proliferator-activated receptor γ coactivator 1α/β (PGC-1) coactivators repress the transcriptional activity of NF-κB in skeletal muscle cells*. J. Biol. Chem. 2012, 288, 2246–2260. [Google Scholar] [CrossRef]
- Chinsomboon, J.; Ruas, J.L.; Gupta, R.K.; Thom, R.; Shoag, J.; Rowe, G.C.; Sawada, N.; Raghuram, S.; Arany, Z. The transcriptional coactivator PGC-1α mediates exercise-induced angiogenesis in skeletal muscle. Proc. Natl. Acad. Sci. USA 2009, 106, 21401–21406. [Google Scholar] [CrossRef]
- Saint-Geniez, M.; Jiang, A.; Abend, S.; Liu, L.; Sweigard, H.; Connor, K.M.; Arany, Z. PGC-1α regulates normal and pathological angiogenesis in the retina. Am. J. Pathol. 2012, 182, 255–265. [Google Scholar] [CrossRef]
- Rowe, G.C.; Jiang, A.; Arany, Z. PGC-1 coactivators in cardiac development and disease. Circ. Res. 2010, 107, 825–838. [Google Scholar] [CrossRef]
- Lai, L.; Leone, T.C.; Zechner, C.; Schaeffer, P.J.; Kelly, S.M.; Flanagan, D.P.; Medeiros, D.M.; Kovacs, A.; Kelly, D.P. Transcriptional coactivators PGC-1 and PGC-l control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008, 22, 1948–1961. [Google Scholar] [CrossRef] [PubMed]
- Martin, O.J.; Lai, L.; Soundarapandian, M.M.; Leone, T.C.; Zorzano, A.; Keller, M.P.; Attie, A.D.; Muoio, D.M.; Kelly, D.P. A role for peroxisome proliferator-activated receptor γ coactivator-1 in the control of mitochondrial dynamics during postnatal cardiac growth. Circ. Res. 2014, 114, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Russell, L.K.; Mansfield, C.M.; Lehman, J.J.; Kovacs, A.; Courtois, M.; Saffitz, J.E.; Medeiros, D.M.; Valencik, M.L.; McDonald, J.A.; Kelly, D.P. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ. Res. 2004, 94, 525–533. [Google Scholar] [CrossRef]
- Arany, Z.; Novikov, M.; Chin, S.; Ma, Y.; Rosenzweig, A.; Spiegelman, B.M. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR- coactivator 1. Proc. Natl. Acad. Sci. USA 2006, 103, 10086–10091. [Google Scholar] [CrossRef] [PubMed]
- Lehman, J.J.; Barger, P.M.; Kovacs, A.; Saffitz, J.E.; Medeiros, D.M.; Kelly, D.P. Peroxisome proliferator–activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Investig. 2000, 106, 847–856. [Google Scholar] [CrossRef]
- Arany, Z.; He, H.; Lin, J.; Hoyer, K.; Handschin, C.; Toka, O.; Ahmad, F.; Matsui, T.; Chin, S.; Wu, P.-H.; et al. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005, 1, 259–271. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, J.; Drori, S.; Uldry, M.; Silvaggi, J.M.; Rhee, J.; Jager, S.; Handschin, C.; Zheng, K.; Lin, J.; Yang, W.; et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 2006, 127, 397–408. [Google Scholar] [CrossRef]
- Blechman, J.; Amir-Zilberstein, L.; Gutnick, A.; Ben-Dor, S.; Levkowitz, G. The metabolic regulator PGC-1 directly controls the expression of the hypothalamic neuropeptide oxytocin. J. Neurosci. 2011, 31, 14835–14840. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, S.; Li, Y.; Yu, S.; Zhao, Y. SIRT1/PGC-1α signaling promotes mitochondrial functional recovery and reduces apoptosis after intracerebral hemorrhage in rats. Front. Mol. Neurosci. 2018, 10, 443. [Google Scholar] [CrossRef]
- Taherzadeh-Fard, E.; Saft, C.; Akkad, D.A.; Wieczorek, S.; Haghikia, A.; Chan, A.; Epplen, J.T.; Arning, L. PGC-1alpha downstream transcription factors NRF-1 and TFAM are genetic modifiers of Huntington disease. Mol. Neurodegener. 2011, 6, 32. [Google Scholar] [CrossRef]
- Soyal, S.M.; Felder, T.K.; Auer, S.; Hahne, P.; Oberkofler, H.; Witting, A.; Paulmichl, M.; Landwehrmeyer, G.B.; Weydt, P.; Patsch, W.; et al. A greatly extended PPARGC1A genomic locus encodes several new brain-specific isoforms and influences Huntington disease age of onset. Hum. Mol. Genet. 2012, 21, 3461–3473. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Redondo, V.; Pettersson, A.T.; Ruas, J.L. The hitchhiker’s guide to PGC-1α isoform structure and biological functions. Diabetologia 2015, 58, 1969–1977. [Google Scholar] [CrossRef] [PubMed]
- Jesse, S.; Bayer, H.; Alupei, M.C.; Zügel, M.; Mulaw, M.A.; Tuorto, F.; Malmsheimer, S.; Singh, K.; Steinacker, J.M.; Schumann, U.; et al. Ribosomal transcription is regulated by PGC-1alpha and disturbed in Huntington’s disease. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Soyal, S.M.; Zara, G.; Ferger, B.; Felder, T.K.; Kwik, M.; Nofziger, C.; Dossena, S.; Schwienbacher, C.; Hicks, A.A.; Pramstaller, P.P.; et al. The PPARGC1A locus and CNS-specific PGC-1α isoforms are associated with Parkinson’s Disease. Neurobiol. Dis. 2019, 121, 34–46. [Google Scholar] [CrossRef]
- Chandra, G.; Shenoi, R.; Anand, R.; Rajamma, U.; Mohanakumar, K. Reinforcing mitochondrial functions in aging brain: An insight into Parkinson’s disease therapeutics. J. Chem. Neuroanat. 2019, 95, 29–42. [Google Scholar] [CrossRef]
- Katsouri, L.; Lim, Y.M.; Blondrath, K.; Eleftheriadou, I.; Lombardero, L.; Birch, A.M.; Mirzaei, N.; Irvine, E.E.; Mazarakis, N.D.; Sastre, M. PPARγ-coactivator-1α gene transfer reduces neuronal loss and amyloid-β generation by reducing β-secretase in an Alzheimer’s disease model. Proc. Natl. Acad. Sci. USA 2016, 113, 12292–12297. [Google Scholar] [CrossRef]
- Sweeney, G.; Song, J. The association between PGC-1α and Alzheimer’s disease. Anat. Cell Biol. 2016, 49, 1–6. [Google Scholar] [CrossRef]
- Mastropasqua, F.; Girolimetti, G.; Shoshan, M.C. PGC1α: Friend or foe in cancer? Genes 2018, 9, 48. [Google Scholar] [CrossRef]
- Bost, F.; Kaminski, L. The metabolic modulator PGC-1α in cancer. Am. J. Cancer Res. 2019, 9, 198–211. [Google Scholar]
- Li, Y.; Wingert, R.A. Regenerative medicine for the kidney: Stem cell prospects & challenges. Clin. Transl. Med. 2013, 2, 11. [Google Scholar] [CrossRef]
- Chambers, J.M.; Poureetezadi, S.J.; Addiego, A.; Lahne, M.; Wingert, R.A. ppargc1a controls nephron segmentation during zebrafish embryonic kidney ontogeny. eLife 2018, 7, e40266. [Google Scholar] [CrossRef] [PubMed]
- Poureetezadi, S.J.; Cheng, C.N.; Chambers, J.M.; Drummond, B.E.; Wingert, R.A. Prostaglandin signaling regulates nephron segment patterning of renal progenitors during zebrafish kidney development. eLife 2016, 5, e17551. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.; Tam, D.; Bardia, A.; Bhasin, M.K.; Rowe, G.C.; Kher, A.; Zsengeller, Z.K.; Akhavan-Sharif, M.R.; Khankin, E.V.; Saintgeniez, M.; et al. PGC-1α promotes recovery after acute kidney injury during systemic inflammation in mice. J. Clin. Investig. 2011, 121, 4003–4014. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2013, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.; Zsengeller, Z.K.; Berg, A.H.; Khankin, E.V.; Bhasin, M.K.; Kim, W.; Clish, C.B.; Stillman, I.E.; Karumanchi, S.A.; Rhee, E.P.; et al. PGC1α drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature 2016, 531, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Casemayou, A.; Fournel, A.; Bagattin, A.; Schanstra, J.P.; Belliere, J.; Decramer, S.; Marsal, D.; Gillet, M.; Chassaing, N.; Huart, A.; et al. Hepatocyte nuclear factor-1β controls mitochondrial respiration in renal tubular cells. J. Am. Soc. Nephrol. 2017, 28, 3205–3217. [Google Scholar] [CrossRef] [PubMed]
- Han, S.H.; Wu, M.-Y.; Nam, B.Y.; Park, J.T.; Yoo, T.-H.; Jhee, J.H.; Park, J.; Chinga, F.; Li, S.-Y.; Susztak, K. PGC-1α protects from notch-induced kidney fibrosis development. J. Am. Soc. Nephrol. 2017, 28, 3312–3322. [Google Scholar] [CrossRef]
- Hasumi, H.; Baba, M.; Hasumi, Y.; Huang, Y.; Oh, H.; Hughes, R.M.; Klein, M.E.; Takikita, S.; Nagashima, K.; Schmidt, L.S.; et al. Regulation of mitochondrial oxidative metabolism by tumor suppressor FLCN. J. Natl. Cancer Inst. 2012, 104, 1750–1764. [Google Scholar] [CrossRef]
- LaGory, E.L.; Wu, C.; Taniguchi, C.M.; Ding, C.-K.C.; Chi, J.-T.; Von Eyben, R.; Scott, D.A.; Richardson, A.D.; Giaccia, A.J. Suppression of PGC-1α is critical for reprogramming oxidative metabolism in renal cell carcinoma. Cell Rep. 2015, 12, 116–127. [Google Scholar] [CrossRef]
- Xiao, W.; Xiong, Z.; Xiong, W.; Yuan, C.; Xiao, H.; Ruan, H.; Song, Z.; Wang, C.; Bao, L.; Cao, Q.; et al. Melatonin/PGC1A/UCP1 promotes tumor slimming and represses tumor progression by initiating autophagy and lipid browning. J. Pineal Res. 2019, 67, e12607. [Google Scholar] [CrossRef]
- Nam, H.; Kundu, A.; Brinkley, G.J.; Chandrashekar, D.S.; Kirkman, R.L.; Chakravarthi, B.V.K.; Orlandella, R.M.; Norian, L.A.; Sonpavde, G.; Ghatalia, P.; et al. PGC1α suppresses kidney cancer progression by inhibiting collagen-induced SNAIL expression. Matrix Biol. 2020, 89, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Felipe-Abrio, B.; Verdugo-Sivianes, E.M.; Carnero, A. c-MYB- and PGC1a-dependent metabolic switch induced by MYBBP1A loss in renal cancer. Mol. Oncol. 2019, 13, 1519–1533. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.-H.; Xu, Y.; Wang, J.; Wan, F.-N.; Wang, H.-K.; Cao, D.-L.; Shi, G.-H.; Qu, Y.-Y.; Zhang, H.-L.; Ye, D.-W. Prognostic value and immune infiltration of novel signatures in clear cell renal cell carcinoma microenvironment. Aging 2019, 11, 6999–7020. [Google Scholar] [CrossRef] [PubMed]
- McCampbell, K.K.; Wingert, R.A. Renal stem cells: Fact or science fiction? Biochem. J. 2012, 444, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Rasbach, K.A.; Schnellmann, R.G. PGC-1α over-expression promotes recovery from mitochondrial dysfunction and cell injury. Biochem. Biophys. Res. Commun. 2007, 355, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Funk, J.A.; Schnellmann, R.G. Accelerated recovery of renal mitochondrial and tubule homeostasis with SIRT1/PGC-1α activation following ischemia–reperfusion injury. Toxicol. Appl. Pharmacol. 2013, 273, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Collier, J.B.; Whitaker, R.M.; Eblen, S.T.; Schnellmann, R.G. Rapid renal regulation of peroxisome proliferator-activated receptor γ coactivator-1α by extracellular signal-regulated kinase 1/2 in physiological and pathological conditions. J. Biol. Chem. 2016, 291, 26850–26859. [Google Scholar] [CrossRef] [PubMed]
- Perry, H.; Huang, L.; Wilson, R.J.; Bajwa, A.; Sesaki, H.; Yan, Z.; Rosin, D.L.; Kashatus, D.F.; Okusa, M.D. Dynamin-related protein 1 deficiency promotes recovery from AKI. J. Am. Soc. Nephrol. 2017, 29, 194–206. [Google Scholar] [CrossRef]
- Fontecha-Barriuso, M.; Martín-Sánchez, D.; Martinez-Moreno, J.M.; Carrasco, S.; Ruiz-Andrés, O.; Monsalve, M.; Sanchez-Ramos, C.; Gómez, M.J.; Ruiz-Ortega, M.; Sánchez-Niño, M.D.; et al. PGC-1α deficiency causes spontaneous kidney inflammation and increases the severity of nephrotoxic AKI. J. Pathol. 2019, 249, 65–78. [Google Scholar] [CrossRef]
- Lim, A.K. Diabetic nephropathy—Complications and treatment. Int. J. Nephrol. Renov. Dis. 2014, 7, 361–381. [Google Scholar] [CrossRef]
- National Institute of Diabetes and Digestive and Kidney Disease NIDDK—National Institute of Health (NIH). Chronic Kidney Disease. 2017. Available online: https://www.niddk.nih.gov/health-information/kidney-disease/chronic-kidney-disease-ckd/what-is-chronic-kidney-disease (accessed on 30 April 2020).
- Sharma, K.; Karl, B.; Mathew, A.V.; Gangoiti, J.A.; Wassel, C.L.; Saito, R.; Pu, M.; Sharma, S.; You, Y.-H.; Wang, L.; et al. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J. Am. Soc. Nephrol. 2013, 24, 1901–1912. [Google Scholar] [CrossRef] [PubMed]
- Dugan, L.L.; You, Y.-H.; Ali, S.S.; Diamond-Stanic, M.; Miyamoto, S.; Declèves, A.-E.; Andreyev, A.Y.; Quach, T.; Ly, S.; Shekhtman, G.; et al. AMPK dysregulation promotes diabetes-related reduction of superoxide and mitochondrial function. J. Clin. Investig. 2013, 123, 4888–4899. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Badal, S.S.; Ye, Z.; Wang, Y.; Ayanga, B.A.; Galvan, D.L.; Green, N.H.; Chang, B.H.; Overbeek, P.A.; Danesh, F.R. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Investig. 2016, 126, 4205–4218. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-Y.; Park, J.; Qiu, C.; Han, S.H.; Palmer, M.B.; Arany, Z.; Susztak, K. Increasing the level of peroxisome proliferator-activated receptor γ coactivator-1α in podocytes results in collapsing glomerulopathy. JCI Insight 2017, 2, e92930. [Google Scholar] [CrossRef] [PubMed]
- Svensson, K.; Schnyder, S.; Cardel, B.; Handschin, C. Loss of renal tubular PGC-1α exacerbates diet-induced renal steatosis and age-related urinary sodium excretion in mice. PLoS ONE 2016, 11, e0158716. [Google Scholar] [CrossRef]
- Kang, H.M.; Ahn, S.H.; Choi, P.; Ko, Y.-A.; Han, S.H.; Chinga, F.; Park, A.S.D.; Tao, J.; Sharma, K.; Pullman, J.; et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2014, 21, 37–46. [Google Scholar] [CrossRef]
- Peng, H.; Wang, Q.; Lou, T.; Qin, J.; Jung, S.; Shetty, V.; Li, F.; Wang, Y.; Feng, X.-H.; Mitch, W.E.; et al. Myokine mediated muscle-kidney crosstalk suppresses metabolic reprogramming and fibrosis in damaged kidneys. Nat. Commun. 2017, 8, 1493. [Google Scholar] [CrossRef]
- Huang, S.; Park, J.; Qiu, C.; Chung, K.W.; Li, S.-Y.; Sirin, Y.; Han, S.H.; Taylor, V.; Zimber-Strobl, U.; Susztak, K. Jagged1/Notch2 controls kidney fibrosis via Tfam-mediated metabolic reprogramming. PLoS Biol. 2018, 16, e2005233. [Google Scholar] [CrossRef]
- National Institute of Diabetes and Digestive and Kidney Disease (NIDDKD). Polycystic Kidney Disease. Available online: https://www.niddk.nih.gov/health-information/kidney-disease/polycystic-kidney-disease (accessed on 30 April 2020).
- Rossetti, S.; Consugar, M.B.; Chapman, A.B.; Torres, V.E.; Guay-Woodford, L.M.; Grantham, J.J.; Bennett, W.M.; Meyers, C.M.; Walker, D.L.; Bae, K.; et al. Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2007, 18, 2143–2160. [Google Scholar] [CrossRef]
- Chapin, H.C.; Caplan, M.J. The cell biology of polycystic kidney disease. J. Cell Biol. 2010, 191, 701–710. [Google Scholar] [CrossRef]
- Qian, F.; Watnick, T.J.; Onuchic, L.F.; Germino, G.G. The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell 1996, 87, 979–987. [Google Scholar] [CrossRef]
- Pei, Y.; Watnick, T.; He, N.; Wang, K.; Liang, Y.; Parfrey, P.; Germino, G.; George-Hyslop, P.S. Somatic PKD2 mutations in individual kidney and liver cysts support a "two-hit" model of cystogenesis in type 2 autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 1999, 10, 1524–1529. [Google Scholar] [PubMed]
- Watnick, T.; He, N.; Wang, K.; Liang, Y.; Parfrey, P.; Hefferton, D.; George-Hyslop, P.S.; Germino, G.G.; Pei, Y. Mutations of PKD1 in ADPKD2 cysts suggest a pathogenic effect of trans-heterozygous mutations. Nat. Genet. 2000, 25, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, Y.; Inagi, R.; Yoshihara, D.; Kugita, M.; Nagao, S.; Shimizu, A.; Takeda, N.; Wake, M.; Honda, K.; Zhou, J.; et al. Mitochondrial abnormality facilitates cyst formation in autosomal dominant polycystic kidney disease. Mol. Cell. Biol. 2017, 37, e00337-17. [Google Scholar] [CrossRef]
- Hajarnis, S.; Lakhia, R.; Yheskel, M.; Williams, D.; Sorourian, M.; Liu, X.; Aboudehen, K.; Zhang, S.; Kersjes, K.; Galasso, R.; et al. microRNA-17 family promotes polycystic kidney disease progression through modulation of mitochondrial metabolism. Nat. Commun. 2017, 8, 14395. [Google Scholar] [CrossRef]
- Li, X. Department of internal medicine epigenetics in ADPKD: Understanding mechanisms and discovering treatment. In Polycystic Kidney Disease; Codon Publications: Singapore, 2015; pp. 283–311. [Google Scholar]
- Chambers, J.M.; Addiego, A.; Wingert, R.A. Ppargc1a controls ciliated cell development by regulating prostaglandin biosynthesis. CELL-REPORTS-D-19-04964. Available online: http://dx.doi.org/10.2139/ssrn.3509910 (accessed on 27 December 2019).
- Gresh, L.; Fischer, E.; Reimann, A.; Tanguy, M.; Garbay, S.; Shao, X.; Hiesberger, T.; Fiette, L.; Igarashi, P.; Yaniv, M.; et al. A transcriptional network in polycystic kidney disease. EMBO J. 2004, 23, 1657–1668. [Google Scholar] [CrossRef]
- Hiesberger, T.; Bai, Y.; Shao, X.; McNally, B.T.; Sinclair, A.M.; Tian, X.; Igarashi, P. Mutation of hepatocyte nuclear factor–1β inhibits Pkhd1 gene expression and produces renal cysts in mice. J. Clin. Invest. 2004, 113, 814–825. [Google Scholar] [CrossRef]
- Jin, D.; Ni, T.T.; Sun, J.; Wan, H.; Amack, J.D.; Yu, G.; Fleming, J.; Chiang, C.; Li, W.; Papierniak, A.; et al. Prostaglandin signalling regulates ciliogenesis by modulating intraflagellar transport. Nat. Cell Biol. 2014, 16, 841–851. [Google Scholar] [CrossRef]
- Marra, A.N.; Adeeb, B.D.; Chambers, B.E.; Drummond, B.E.; Ulrich, M.; Addiego, A.; Springer, M.; Poureetezadi, S.J.; Chambers, J.M.; Ronshaugen, M.R.; et al. Prostaglandin signaling regulates renal multiciliated cell specification and maturation. Proc. Natl. Acad. Sci. USA 2019, 116, 8409–8418. [Google Scholar] [CrossRef]
- Boehlke, C.; Kotsis, F.; Patel, V.; Braeg, S.; Voelker, H.; Bredt, S.; Beyer, T.; Janusch, H.; Hamann, C.; Gödel, M.; et al. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat. Cell Biol. 2010, 12, 1115–1122. [Google Scholar] [CrossRef]
- Burkhalter, M.D.; Sridhar, A.; Sampaio, P.; Jacinto, R.; Burczyk, M.S.; Donow, C.; Angenendt, M.; Hempel, M.; Walther, P.; Pennekamp, P.; et al. Imbalanced mitochondrial function provokes heterotaxy via aberrant ciliogenesis. J. Clin. Investig. 2019, 129, 2841–2855. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, J.F.; Liu, Y.; Davis, E.E.; Westlake, C.J.; Attanasio, M.; Otto, E.A.; Seelow, D.; Nurnberg, G.; Becker, C.; Nuutinen, M.; et al. Individuals with mutations in XPNPEP3, which encodes a mitochondrial protein, develop a nephronophthisis-like nephropathy. J. Clin. Investig. 2010, 120, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Kang, S.U.; Zhang, S.; Karuppagounder, S.; Xu, J.; Lee, Y.-K.; Kang, B.-G.; Lee, Y.; Zhang, J.; Pletnikova, O.; et al. Adult conditional knockout of PGC-1α leads to loss of dopamine neurons. eNeuro 2016, 3, ENEURO.0183-16.2016. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chambers, J.M.; Wingert, R.A. PGC-1α in Disease: Recent Renal Insights into a Versatile Metabolic Regulator. Cells 2020, 9, 2234. https://doi.org/10.3390/cells9102234
Chambers JM, Wingert RA. PGC-1α in Disease: Recent Renal Insights into a Versatile Metabolic Regulator. Cells. 2020; 9(10):2234. https://doi.org/10.3390/cells9102234
Chicago/Turabian StyleChambers, Joseph M., and Rebecca A. Wingert. 2020. "PGC-1α in Disease: Recent Renal Insights into a Versatile Metabolic Regulator" Cells 9, no. 10: 2234. https://doi.org/10.3390/cells9102234
APA StyleChambers, J. M., & Wingert, R. A. (2020). PGC-1α in Disease: Recent Renal Insights into a Versatile Metabolic Regulator. Cells, 9(10), 2234. https://doi.org/10.3390/cells9102234





