Journal Description
Epigenomes
Epigenomes
is an international, peer-reviewed, open access journal on epigenetics and epigenomics, published quarterly online by MDPI. The Epigenetics Society is affiliated with Epigenomes and its members receive discounts on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, ESCI (Web of Science), PMC, PubMed, Embase, PubAg, CAPlus / SciFinder, and other databases.
- Journal Rank: JCR - Q2 (Genetics and Heredity) / CiteScore - Q2 (Biochemistry, Genetics and Molecular Biology (miscellaneous))
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 25.5 days after submission; acceptance to publication is undertaken in 3.8 days (median values for papers published in this journal in the second half of 2025).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
3.5 (2024);
5-Year Impact Factor:
3.1 (2024)
Latest Articles
Epigenetic Liquid Biopsy Marks Atrial Fibrillation: Evidence from the AF Big Picture Study
Epigenomes 2026, 10(1), 9; https://doi.org/10.3390/epigenomes10010009 - 5 Feb 2026
Abstract
Background/Objectives: Atrial fibrillation (AF) is currently the most common arrhythmia worldwide, and it is linked to increased mortality and morbidity, hence the need for a better clinical stratification of AF patients. Histone complexes or nucleosomes, released into the blood circulation, are found
[...] Read more.
Background/Objectives: Atrial fibrillation (AF) is currently the most common arrhythmia worldwide, and it is linked to increased mortality and morbidity, hence the need for a better clinical stratification of AF patients. Histone complexes or nucleosomes, released into the blood circulation, are found elevated in acute conditions such as stroke, trauma, and sepsis. The aim of this pilot single-centre study was to assess whether circulating histone levels could be used for diagnostic purposes in patients with AF. Methods: A total of 40 patients, well characterised for their biochemical and clinical characteristics, were recruited from outpatient clinics. Patients were randomly recruited into two groups (n = 20 per group), i.e., persistent AF and hypertensive controls. A multi-channel flow imaging methodology based on ImageStreamX was used with a well-optimised protocol to image and quantify five individual histones (H2A, H2B, H3, H4, and macroH2A1.1) together with the dimers (H2A/H2B, and H3/H4). Results: In the AF groups, plasma levels of histone dimers H2A/H2B and H3/H4 were elevated compared to hypertensive controls, 1.8% vs. 1.06% (p-value = 0.03). H2A/H2B dimer levels were increased in AF patients irrespective of gender, smoking status, diabetes, and pharmacological therapy. In the overall population, an inverse correlation between H2A and BMI was detected. Conclusions: Our pilot study, although limited in sample size, suggests that circulating histone complexes may be epigenetic sentinels for AF, offering mechanistic insights while addressing unmet needs in risk stratification.
Full article
(This article belongs to the Special Issue Epigenetic Signatures in Metabolic Health and Cancer)
►
Show Figures
Open AccessArticle
Differences in MicroRNA Expression in Firefighters Responding to a Train Derailment and Fire in East Palestine, Ohio
by
Jaclyn M. Goodrich, Yaodong Xin, Shawn C. Beitel, John Gulotta, Lu Wang, Bhavya Thotakura, Judith M. Graber, Derek Urwin, Alexander C. Mayer, Sara Jahnke, Derrick L. Edwards, Casey Grant, Sreenivasan Ranganathan and Jefferey L. Burgess
Epigenomes 2026, 10(1), 8; https://doi.org/10.3390/epigenomes10010008 - 3 Feb 2026
Abstract
►▼
Show Figures
Background/Objectives: High-risk, low-frequency incidents such as building collapses and large chemical fires can result in acute, high-dose exposures to toxic agents for first responders and the surrounding community. While these exposures may last for hours to days, their contribution to firefighters’ risks
[...] Read more.
Background/Objectives: High-risk, low-frequency incidents such as building collapses and large chemical fires can result in acute, high-dose exposures to toxic agents for first responders and the surrounding community. While these exposures may last for hours to days, their contribution to firefighters’ risks for cancer and other diseases is relatively unknown. In February 2023, a freight train transporting chemicals derailed and caught fire in East Palestine, Ohio, US. More than 350 firefighters, primarily volunteer, responded to the incident. In this cross-sectional study, we evaluated epigenetic markers of toxicity in responding firefighters. We hypothesized that exposures from responding to the train derailment would alter the expression of microRNAs (miRNAs) linked to carcinogenesis. Methods: We enrolled 62 responding firefighters and a comparison group of 26 firefighters from the same region who did not respond to the incident. We measured the relative expression of 800 miRNAs in blood samples using the nCounter Human v3 miRNA expression panel. We compared the expression of miRNA between exposure groups in negative binomial regression models, adjusting for potential confounders. Results: At a false discover rate cut-off of 5% (q-value < 0.05), 16 miRNAs had significantly higher expression and one significantly lower among firefighters that responded to the incident. Top disease-related pathways in which these miRNAs were enriched included those relevant to neurodegenerative diseases, vascular disease, and multiple cancer sites. Conclusions: Overall, results suggest responding to one large incident can have non-transient impacts on miRNA expression. Whether this translates into longer-term health risks or adaptive responses to exposures is unclear.
Full article

Figure 1
Open AccessArticle
An Hsp70 Chaperone Is Involved in Meiotic Silencing by Unpaired DNA
by
Victor T. Sy, Sterling S. Trawick, Hagen M. Tatarsky and Patrick K. T. Shiu
Epigenomes 2026, 10(1), 7; https://doi.org/10.3390/epigenomes10010007 - 26 Jan 2026
Abstract
In the filamentous fungus Neurospora crassa, a gene not having a pairing partner during meiosis is seen as a potential intruder and is targeted by a mechanism called meiotic silencing by unpaired DNA (MSUD). MSUD employs core RNA interference (RNAi) components such
[...] Read more.
In the filamentous fungus Neurospora crassa, a gene not having a pairing partner during meiosis is seen as a potential intruder and is targeted by a mechanism called meiotic silencing by unpaired DNA (MSUD). MSUD employs core RNA interference (RNAi) components such as the SMS-2 Argonaute, which uses small interfering RNAs (siRNAs) as guides to seek out mRNAs from unpaired genes for silencing. In Drosophila melanogaster, the heat shock protein 70 (Hsp70) chaperone system facilitates the conformational activation of an Argonaute and allows it to load siRNAs. Here, our results demonstrate that an Hsp70 protein in Neurospora interacts with SMS-2 and mediates the silencing of unpaired genes.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures

Figure 1
Open AccessArticle
ARS2, a Cofactor of CBC, Promotes Meiotic Silencing by Unpaired DNA
by
Michael M. Vierling, Victor T. Sy, Logan M. Decker, Hua Xiao, Justine N. Hemaya and Patrick K. T. Shiu
Epigenomes 2026, 10(1), 6; https://doi.org/10.3390/epigenomes10010006 - 21 Jan 2026
Abstract
The presence of an extra DNA segment in a genome could indicate a transposon or another repetitive element on the move. In Neurospora crassa, a surveillance mechanism called meiotic silencing by unpaired DNA (MSUD) is maintained to monitor these selfish elements. MSUD
[...] Read more.
The presence of an extra DNA segment in a genome could indicate a transposon or another repetitive element on the move. In Neurospora crassa, a surveillance mechanism called meiotic silencing by unpaired DNA (MSUD) is maintained to monitor these selfish elements. MSUD utilizes common RNA interference (RNAi) factors, including the SMS-2 Argonaute, to target mRNAs from genes lacking a pairing partner during meiosis. In eukaryotes, an mRNA transcript is typically bound at the 5′ cap by the cap-binding complex (CBC), which assists in its nuclear export. Previously, we discovered that CBC and its interactor NCBP3 mediate MSUD, possibly by guiding the perinuclear SMS-2 to effectively recognize exported mRNAs. Here, we report that ARS2, a CBC cofactor, is involved in MSUD. ARS2 interacts with both CBC and NCBP3, and it may help bring them together. In addition to its role in silencing, ARS2 also contributes to vegetative growth and sexual sporulation.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures

Figure 1
Open AccessReview
Something Old, Something New, Something Borrowed… About the Placenta
by
Nadezhda Milova, Maria Nikolova, Angel Yordanov, Antoan Milov and Stoilka Mandadzhieva
Epigenomes 2026, 10(1), 5; https://doi.org/10.3390/epigenomes10010005 - 19 Jan 2026
Abstract
The connection between the mother and the child has been considered one of the strongest bonds in nature. Though there are numerous factors that can influence the establishment of pregnancy, in its essence, three are considered major: a good quality embryo, a receptive
[...] Read more.
The connection between the mother and the child has been considered one of the strongest bonds in nature. Though there are numerous factors that can influence the establishment of pregnancy, in its essence, three are considered major: a good quality embryo, a receptive endometrium, and successful cross-talk between them. The placenta, which derives from the trophoblast of the embryo, develops when a successful implantation occurs. It is an ephemeral organ through which the turnover of nutrients, gases, and waste molecules is realized. It serves as a barrier and can provide the embryo with immune factors. Placental disorders are observed in some rare but life-threatening obstetric conditions like preeclampsia (PE), fetal growth restriction (FGR), gestational trophoblastic diseases (GTDs), and gestational diabetes mellitus (GDM). The etiology and pathogenesis of some are still partially enigmatic. Our attention in this review was driven by the participation of small RNA molecules—miRNAs and piRNAs—as potential epigenetic modulators of genes that play a pivotal role in placental functioning. In this study, we analyze the influence of these epigenetic factors on the mechanisms of the development of preeclampsia. The molecular approach for understanding placental disorders may help new diagnostic and therapeutic solutions to be found.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures

Figure 1
Open AccessReview
Epigenetic Alterations in Colitis-Associated Colorectal Cancer
by
Nisha Ganesh, William M. Grady and Andrew M. Kaz
Epigenomes 2026, 10(1), 4; https://doi.org/10.3390/epigenomes10010004 - 16 Jan 2026
Abstract
Colitis-associated colorectal cancer (CAC) represents a distinct subtype of colorectal malignancy that arises in the setting of chronic inflammatory bowel disease (IBD). Unlike sporadic colorectal cancer, CAC develops through inflammation-driven molecular pathways, in which epigenetic alterations play a pivotal role in tumor initiation
[...] Read more.
Colitis-associated colorectal cancer (CAC) represents a distinct subtype of colorectal malignancy that arises in the setting of chronic inflammatory bowel disease (IBD). Unlike sporadic colorectal cancer, CAC develops through inflammation-driven molecular pathways, in which epigenetic alterations play a pivotal role in tumor initiation and progression. This review highlights the major epigenetic mechanisms implicated in CAC, including DNA methylation, histone modifications, and microRNA (miRNA) dysregulation. Aberrant DNA methylation patterns, such as promoter hypermethylation of tumor suppressor genes and global hypomethylation, contribute to genomic instability and altered gene expression. In parallel, inflammation-induced changes in histone configuration modulate chromatin accessibility and transcriptional activity of key oncogenic and tumor-suppressive pathways. Furthermore, deregulated miRNAs influence multiple aspects of CAC pathogenesis by targeting genes involved in inflammation and tumor progression. Understanding these epigenetic processes provides valuable insights into the development of colorectal malignancy and identifies potential biomarkers for early detection and intervention in colitis-associated colorectal cancer.
Full article
(This article belongs to the Special Issue Epigenetic Signatures in Metabolic Health and Cancer)
Open AccessArticle
Fisetin Suppresses the Proliferative and Migratory Behavior of HeLa Cells by Modulating Aberrant Epigenetic Marks (Writers and Erasers)
by
Nazia Afroze, Reham I. Alagal, Lujain A. Almousa, Ritu Raina, Prathap Bava, Lizna Mohamed Ali, Tarique Noorul Hasan and Arif Hussain
Epigenomes 2026, 10(1), 3; https://doi.org/10.3390/epigenomes10010003 - 12 Jan 2026
Abstract
Purpose: The reversible deviant in epigenomic modulations is the highlight of developing new anti-cancer drugs, necessitating the use of fisetin as an epigenetic modifier in the study. Methods: In silico and molecular studies were performed to analyze the modulatory effect of fisetin on
[...] Read more.
Purpose: The reversible deviant in epigenomic modulations is the highlight of developing new anti-cancer drugs, necessitating the use of fisetin as an epigenetic modifier in the study. Methods: In silico and molecular studies were performed to analyze the modulatory effect of fisetin on various writers and erasers. Further, whole genome DNA methylation sequencing and expression studies were performed. Global DNA methylation-LINE 1 kit was used to check global DNA methylation. Additionally, the effect of fisetin on migration was evaluated by colony, scratch, and invasion assays and qPCR and protein expression studies of migration-related genes were carried out on HeLa cells. Results: In silico studies have supported that fisetin interacts with writers and erasers in their catalytic site and the simulation studies showed minimum fluctuations in energy and temperature over a 10 ns timescale indicating that these complexes are likely to remain stable. Fisetin (20–50 µM) dose-dependently inhibited DNA methyltransferases (DNMT), histone deacetyl transferases (HDAC), histone acetyl transferases (HAT), and histone methyltransferases (HMT) activities at 48 h, with inhibition ranging from 24 to 72% compared to the control. The expression and enzymatic activity of these proteins, along with various H4 and H3 modification marks, were observed to be altered following fisetin treatment at 48 h. Fisetin treatment reduced promoter methylation in various tumor suppressor genes ranging from 15.29% to 76.23% and leading to the corresponding reactivation of important tumor suppressor genes; however, it did not lead to any alteration in the global DNA methylation compared to untreated controls linked with the anti-migratory properties of fisetin as the percentage of migrated cells dropped from ~40% to ~8%. Conclusions: This study gives a mechanistic insight of fisetin as a potential epigenetic modifier in HeLa cells.
Full article
(This article belongs to the Collection Epigenetic Regulation of Cellular Differentiation)
►▼
Show Figures

Figure 1
Open AccessArticle
Epigenetic Regulation and Gene Expression Profiles in Cervical Swabs: Toward Non-Invasive Biomarkers of Cervical Lesion Progression
by
Ivana Kašubová, Andrea Hornáková, Lucia Kotúľová, Tomáš Rokos, Zuzana Kolková, Andrea Kapinová, Terézia Pribulová, Erik Kozubík, Michal Kalman, Kamil Biringer, Erik Kúdela and Veronika Holubeková
Epigenomes 2026, 10(1), 2; https://doi.org/10.3390/epigenomes10010002 - 7 Jan 2026
Abstract
Background/Objectives: Cervical cancer is a common malignancy in women worldwide, closely associated with persistent human papillomavirus (HPV) infection. Epigenetic mechanisms, particularly promoter methylation, may contribute to tumour progression. This pilot study aimed to analyse the promoter methylation patterns and gene expression of
[...] Read more.
Background/Objectives: Cervical cancer is a common malignancy in women worldwide, closely associated with persistent human papillomavirus (HPV) infection. Epigenetic mechanisms, particularly promoter methylation, may contribute to tumour progression. This pilot study aimed to analyse the promoter methylation patterns and gene expression of selected genes (DNMT, BCL2, CDH1, CD8A, MUC1, ALCAM). The goal was to identify associations between promoter hypermethylation, gene expression, and HPV infection in cervical swab specimens obtained from patients with low-grade squamous intraepithelial lesions (SILs), high-grade SILs, or squamous cell carcinomas. Methods: A total of 81 cervical swab samples from Slovak participants were included in the study. DNA methylation and gene expression profiling was performed using real-time PCR (qPCR) and pyrosequencing. Results: BCL2 expression was significantly reduced across all lesion grades. CD8A expression was slightly elevated in low- and high-grade SILs, particularly in HPV-positive samples. MUC1 showed variability with lesion grade. No statistically significant differences in DNA methylation were observed across groups stratified by HPV status, community state type, and lesion grade. Conclusions: Our findings suggest that BCL2 downregulation and gene activity variability influenced by the vaginal microbiome may play a role in cervical lesion progression. These results highlight potential non-invasive biomarkers for monitoring cervical lesions.
Full article
(This article belongs to the Special Issue Epigenetic Signatures in Metabolic Health and Cancer)
►▼
Show Figures

Figure 1
Open AccessArticle
Upregulation of a MicroRNA Signature Involving miR-17-5p, miR-26b-5p, miR-106a-5p, and miR-146a-5p During Cervical Epithelial Transformation
by
Andrea Hornakova, Zuzana Kolkova, Lucia Kotulova, Tomas Rokos, Ivana Kasubova, Terezia Pribulova, Erik Kozubik, Kamil Biringer, Erik Kudela and Veronika Holubekova
Epigenomes 2026, 10(1), 1; https://doi.org/10.3390/epigenomes10010001 - 26 Dec 2025
Abstract
Background: Cervical cancer remains the fourth most common malignancy among women worldwide. Despite vaccination and regular screening, new molecular biomarkers are needed for improved early detection and risk assessment. MicroRNAs (miRNAs) play crucial roles in post-transcriptional regulation, and their dysregulation may contribute
[...] Read more.
Background: Cervical cancer remains the fourth most common malignancy among women worldwide. Despite vaccination and regular screening, new molecular biomarkers are needed for improved early detection and risk assessment. MicroRNAs (miRNAs) play crucial roles in post-transcriptional regulation, and their dysregulation may contribute to cervical carcinogenesis. This study evaluated the expression of selected miRNAs in cervical swab samples and corresponding biopsies from women with various grades of cervical lesions and assessed their relationship with human papillomavirus (HPV) infection. Methods: A total of 72 cervical swab samples were included in this study, divided according to cytological severity: negative for intraepithelial lesion or malignancy (NILM, n = 15), atypical squamous cells of undetermined significance (ASC-US, n = 12), low-grade squamous intraepithelial lesion (LSIL, n = 19), and high-grade squamous intraepithelial lesion (HSIL, n = 26). In a subset of patients, corresponding biopsy specimens were analysed for comparison. The association of miRNA expression with HPV infection status was also examined. miRNA expression was quantified by real-time PCR using commercially available assays. Results: To assess the relationship between miRNA expression, lesion severity, and HPV infection, fold change values were compared to the control group (NILM). No significant differences were observed in the ASC-US group (p > 0.05). In contrast, several miRNAs were significantly upregulated in the LSIL and/or HSIL groups, as well as in HPV-positive samples, indicating their association with both lesion progression and viral infection. Specifically, miR-17-5p, miR-26b-5p, miR-29a-3p, miR-103a-3p, miR-106a-5p, miR-146a-5p, miR-155-5p, and miR-191-5p showed increased expression (p < 0.05) compared with controls. The observed upregulation of miR-26b-5p, miR-106a-5p, and miR-146a-5p highlights their potential role in HPV-associated cervical carcinogenesis. Dysregulated miRNAs were enriched in pathways related to infectious diseases, various types of cancer, and cell adhesion processes. Conclusions: The gradual increase in specific miRNAs with lesion severity and HPV infection suggests their role in cervical carcinogenesis. The identified miRNAs may serve as promising non-invasive biomarkers for early detection and monitoring of HPV-associated cervical lesions.
Full article
(This article belongs to the Special Issue Epigenetic Signatures in Metabolic Health and Cancer)
►▼
Show Figures

Figure 1
Open AccessArticle
Epigenome-Wide Search for Distinctive Methylation Biomarkers of Endothelial and Leukocyte DNA
by
Valeria A. Korolenya, Maxim L. Filipenko and Mariya A. Smetanina
Epigenomes 2025, 9(4), 53; https://doi.org/10.3390/epigenomes9040053 - 17 Dec 2025
Abstract
►▼
Show Figures
The endothelium, as the inner layer of the vascular wall, is in constant contact with blood components, so that leukocytes have the ability to adhere to endotheliocytes and penetrate to the subendothelial space. When studying heterogenic vascular samples containing endothelial cells or pathological
[...] Read more.
The endothelium, as the inner layer of the vascular wall, is in constant contact with blood components, so that leukocytes have the ability to adhere to endotheliocytes and penetrate to the subendothelial space. When studying heterogenic vascular samples containing endothelial cells or pathological processes related to inflammation within the endothelium, it may be necessary to distinguish DNA by endothelial and leukocyte origin, which is possible due to its specific epigenetic modifications. To identify CpG loci that could serve as markers for endothelial cells, we searched for their distinctive stable methylated or demethylated states by applying marginal filtering (selecting CpG loci with methylation Beta values closer to 0 and 1) to the microarray data and identified 47 CpG loci with relatively stable methylation/demethylation status that differentiate endothelial (HUVEC, HCMEC, HPAEC, HPMEC, and LSEC) DNA from leukocyte (granulocytes, monocytes, and lymphocytes) DNA. In addition, we compared CpG loci with high and low levels of DNA methylation between different types of endothelial cells and leukocytes. We believe that the obtained data will hopefully facilitate further studies on endothelial dysfunction.
Full article
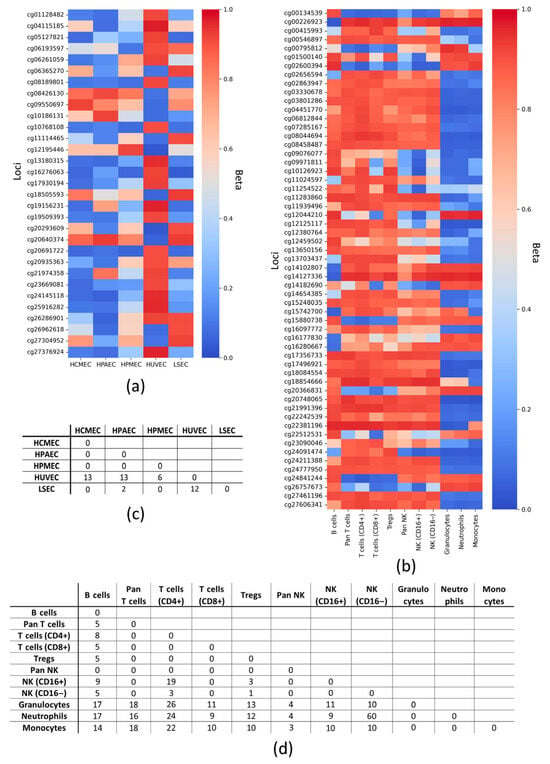
Figure 1
Open AccessArticle
The Exosome-Mediated Epigenome: Non-Coding RNA and mRNA-Coding Networks in Microbiome–Cellular Communication, Inflammation, and Tumorigenesis Along the Oral–Gut–Lung Axis
by
Beatriz Andrea Otálora-Otálora, César Payán-Gómez, Juan Javier López-Rivera, Luisa Fernanda Patiño-Unibio, Sally Lorena Arboleda-Mojica, Claudia Aristizábal-Guzmán, Mario Arturo Isaza-Ruget and Carlos Arturo Álvarez-Moreno
Epigenomes 2025, 9(4), 52; https://doi.org/10.3390/epigenomes9040052 - 16 Dec 2025
Abstract
Background/Objectives: The oral–gut–lung axis represents a dynamic system where exosomes carrying mRNAs and non-coding RNAs might help to regulate microbiota and human cell crosstalk to establish transcriptional regulatory networks controlling cellular biological processes and signaling pathways. Methods: We conducted a comprehensive
[...] Read more.
Background/Objectives: The oral–gut–lung axis represents a dynamic system where exosomes carrying mRNAs and non-coding RNAs might help to regulate microbiota and human cell crosstalk to establish transcriptional regulatory networks controlling cellular biological processes and signaling pathways. Methods: We conducted a comprehensive transcriptomic analysis to characterize the molecular cargo of extracellular exosomes in the context of gut and lung cancer. Results: By analyzing gut and lung exosomes cargo with our previous transcriptomic studies from tumoral and inflammatory tissues, we found that exosomes can transport key RNAs that codify specific receptors that facilitate pathogenic interaction with microorganisms and RNAs that are part of interacting gene and transcriptional regulatory networks that control the function of differentially expresses genes, all involved in biological processes like cell cycle, plasticity and growth regulation, invasion, metastasis, microenvironmental remodeling, epigenetic, and microbial and immunological modulation, during the unlocking of phenotypic plasticity for the acquisition of the hallmarks of cancer in the oral–gut–lung axis. Conclusions: Exosomal RNA regulation of transcriptional networks represents a pivotal axis in the interplay between inflammation and cancer, offering opportunities for innovative diagnostic and therapeutic approaches.
Full article
(This article belongs to the Special Issue Features Papers in Epigenomes 2025)
►▼
Show Figures

Figure 1
Open AccessReview
The Evolutionary Misfit: Evolution, Epigenetics, and the Rise of Non-Communicable Diseases
by
Stefano Amatori
Epigenomes 2025, 9(4), 51; https://doi.org/10.3390/epigenomes9040051 - 13 Dec 2025
Abstract
Human life expectancy has risen dramatically in the last century, but this demographic triumph has come at the cost of an explosion of non-communicable diseases (NCDs), threatening the sustainability of healthcare systems in aging, low-fertility societies. Evolutionary medicine provides a framework to understand,
[...] Read more.
Human life expectancy has risen dramatically in the last century, but this demographic triumph has come at the cost of an explosion of non-communicable diseases (NCDs), threatening the sustainability of healthcare systems in aging, low-fertility societies. Evolutionary medicine provides a framework to understand, at least in part, this paradox. Many vulnerabilities to disease are not failures of design but the predictable outcomes of evolutionary trade-offs, constraints, and mismatches. Evolutionary mismatch theory explains how traits once advantageous in ancestral environments become maladaptive in modern contexts of abundance, sedentarism, and urbanization. The developmental origins of health and disease (DOHaD) concept describes how epigenetic plasticity in early life can buffer or amplify these mismatches, depending on whether adult environments align with developmental forecasts. Transgenerational epigenetic inheritance, even if still debated in humans, may further influence phenotypic plasticity, increasing or mitigating the mismatch. In evolutionary terms, the theories of mutation accumulation, antagonistic pleiotropy, and the disposable soma explain why longer lifespans, and ecological and social conditions profoundly different from those in which we developed, increase the likelihood that these costs are expressed clinically. Because most NCDs can be prevented and effectively controlled but not cured, efforts should prioritize quality of life for people, families, and communities. At the individual level, aligning lifestyles with evolved biology can mitigate risk, but the greatest leverage lies in population-level interventions. Urban health strategies represent a forward-looking attempt to realign modern environments with human biology. In this way, the concept of the evolutionary misfit becomes not just a diagnosis of maladaptation, but a guide for building healthier, more sustainable societies.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures

Figure 1
Open AccessReview
The Epigenomic Impact of Quantum Dots: Emerging Biosensors and Potential Disruptors
by
Abhishu Chand and Kyoungtae Kim
Epigenomes 2025, 9(4), 50; https://doi.org/10.3390/epigenomes9040050 - 8 Dec 2025
Abstract
Quantum dots (QDs) have emerged as powerful tools in biomedical applications due to their unique optical and fluorescent properties which enable highly sensitive and multiplexed detection of biomolecules. Particularly in the field of epigenetic research, QDs are utilized as biosensors for monitoring changes
[...] Read more.
Quantum dots (QDs) have emerged as powerful tools in biomedical applications due to their unique optical and fluorescent properties which enable highly sensitive and multiplexed detection of biomolecules. Particularly in the field of epigenetic research, QDs are utilized as biosensors for monitoring changes in DNA methylation, microRNA (miRNA) expression, and histone modifications, providing a viable alternative to conventional assays. However, increasing evidence also suggests that QDs act as an epigenetic disruptor, altering epigenetic mechanisms and downstream cellular processes. This dual role raises important questions about the safety, reliability, and translational potential of QDs in clinical usage. Therefore, in this commentary we critically evaluate the advances of QD-based epigenetic sensing platforms while also providing insights into QD-based epigenetic dysregulation. We further discuss the current limitations and provide future directions to gain a better understanding of how QDs function to bridge the gap between their diagnostic potential and clinical integration.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures
Figure 1
Open AccessArticle
Introducing the EpG2 System: Epigenomic Processes and the Emergent Genome
by
Edward A. Ruiz-Narváez
Epigenomes 2025, 9(4), 49; https://doi.org/10.3390/epigenomes9040049 - 5 Dec 2025
Abstract
Background/Objectives: Current genomics research equates the genome with DNA sequence and treats the epigenome as a regulatory layer. This DNA-centric view obscures the fact that genomic identity arises through epigenomic processes. The objective of this article is to reinterpret published findings into a
[...] Read more.
Background/Objectives: Current genomics research equates the genome with DNA sequence and treats the epigenome as a regulatory layer. This DNA-centric view obscures the fact that genomic identity arises through epigenomic processes. The objective of this article is to reinterpret published findings into a new theoretical framework: the EpG2 (Epigenome–Genome) system. Methods: This work develops a new conceptual framework by integrating published evidence from diverse domains—including enhancer biology, overlapping genomic functions, alternative coding frames, zygotic genome activation, and disease-associated loci—and reinterpreting these findings through the lens of epigenomic processes. Results: Evidence shows that enhancers emerge only through the interplay of sequence, transcription factors, and chromatin environment. At fertilization, paternal and maternal genomes remain separate, and a new genome emerges through coordinated epigenomic reprogramming or zygote genome emergence (ZGE). DNA sequence risk variants illustrate the concept of contextual risk alleles, whose effects shift across tissues and developmental stages as epigenomic contexts change. Conclusions: The EpG2 system reframes the genome as a processual, emergent entity generated and regulated by epigenomic processes, offering a paradigm for understanding genomic variation beyond DNA sequence.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures

Figure 1
Open AccessArticle
Adverse Childhood Experiences, DNA Methylation, and Depressive Symptoms in Black Pregnant Women
by
Alexandra L. Nowak, Marvin A. Schilt-Solberg, Xiaoyu Liang, Fabiola Magaña, Dawn P. Misra and Carmen Giurgescu
Epigenomes 2025, 9(4), 48; https://doi.org/10.3390/epigenomes9040048 - 27 Nov 2025
Abstract
Background: Prenatal depression, affecting up to a quarter of all pregnancies in the United States, contributes to morbidity and mortality and is associated with increased risk of adverse birth and long-term mental health outcomes. Adverse childhood experiences (ACEs, or experiences of abuse, neglect,
[...] Read more.
Background: Prenatal depression, affecting up to a quarter of all pregnancies in the United States, contributes to morbidity and mortality and is associated with increased risk of adverse birth and long-term mental health outcomes. Adverse childhood experiences (ACEs, or experiences of abuse, neglect, or family dysfunction experienced prior to age 18) are a strong predictor of adult depression and adverse health outcomes. The present study investigated whether epigenetic modification in the form of DNA methylation (DNAm) of four stress-related, glucocorticoid pathway genes (CRH, CRHR1, FKBP5, NR3C1) mediates associations between ACEs and depressive symptoms among Black pregnant women. Methods: Using a cross-sectional design, we examined the mediating role of DNAm on the relationship between depressive symptoms (Center for Epidemiologic Studies Depression Scale (CES-D)) and ACEs (Centers for Disease Control and Prevention 10-item questionnaire), in a subsample (n = 61) of Black pregnant women who were participants of the Biosocial Impacts of Black Births (BIBB) study. Results: A significant association was found between ACEs and depressive symptoms scores (TE α_X = 2.29 with p_TE = 6.60 × 105). DNAm on five CpG sites within two genes significantly mediated the relationship between ACEs and depressive symptoms (cg03238273 on CRHR1, and cg08845721, cg16594263, cg19820298, and cg23430507 on NR3C1). Conclusions: This study provides evidence that DNAm partially mediated the association of ACEs and depressive symptoms during pregnancy among Black pregnant women. Understanding the molecular pathways underlying the mediating effect of ACEs on depressive symptoms among Black pregnant women can illuminate biological markers that help identify and treat pregnant women who are at an increased risk for depression following childhood trauma.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures

Figure 1
Open AccessReview
CircRNAs—Potential Diagnostic Biomarkers and Therapeutic Targets for Receptive and Cancerous Endometrium
by
Antoan Milov, Maria Nikolova, Stoilka Mandadzhieva, Nina Doncheva, Nadezhda Milova and Angel Yordanov
Epigenomes 2025, 9(4), 47; https://doi.org/10.3390/epigenomes9040047 - 17 Nov 2025
Abstract
Circular RNAs (circRNAs) are small, non-coding RNAs in which the 5′ and 3′ ends are linked covalently by back-splicing of exons from a single pre-mRNA. More and more scientific evidence is gathered for their wide distribution in the animal world, playing the role
[...] Read more.
Circular RNAs (circRNAs) are small, non-coding RNAs in which the 5′ and 3′ ends are linked covalently by back-splicing of exons from a single pre-mRNA. More and more scientific evidence is gathered for their wide distribution in the animal world, playing the role of regulators for biological processes, being cell- and tissue-specific. They can influence cellular physiology by various molecular mechanisms, finally modulating gene expression. CircRNAs are believed nowadays to be expressed in both receptive and cancerous endometrium. Due to their abundant expression in the endometrial tissue and their small size and stability, they have been considered potential diagnostic markers and treatment targets for endometrial-related diseases. The regulation of proliferation and differentiation is essential for the formation of receptive endometrium and for endometrial cancer emergence and progression. The receptive endometrium can be regarded as the most highly differentiated state of the endometrium. In contrast, the cancerous endometrium is characterized by a high level of proliferation and the lowest degree of differentiation. These endometria could be conditionally considered opposites. We are investigating the circRNA–miRNA–mRNA regulatory networks that can promote or suppress the proliferation and differentiation of endometrial cells by activating specific signaling pathways in both receptive and cancerous endometria. It could be worth knowing whether there are universal endometrial switches responsible for proliferation and differentiation processes that can alter the balance between them. We are interested in their clinical application as biomarkers and therapeutic targets for both endometrial receptivity issues and EC cases, particularly in diagnosis, progression assessment, and outcome prediction.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures

Figure 1
Open AccessReview
Epigenetic Regulation of Salt Stress Responses in Rice: Mechanisms and Prospects for Enhancing Tolerance
by
Emanuela Talarico, Eleonora Greco, Francesco Guarasci, Fabrizio Araniti, Adriana Chiappetta and Leonardo Bruno
Epigenomes 2025, 9(4), 46; https://doi.org/10.3390/epigenomes9040046 - 16 Nov 2025
Cited by 1
Abstract
►▼
Show Figures
Rice (Oryza sativa L.) is a staple food for over half the global population and a model organism for monocot plant research. However, it is susceptible to salinity, with most cultivated varieties showing reduced growth at salt levels above 3 dS/m. Despite
[...] Read more.
Rice (Oryza sativa L.) is a staple food for over half the global population and a model organism for monocot plant research. However, it is susceptible to salinity, with most cultivated varieties showing reduced growth at salt levels above 3 dS/m. Despite numerous efforts to improve its salt tolerance, little progress has been made. A promising area of research lies in the study of epigenetic regulation, which encompasses DNA methylation, histone modifications, and chromatin remodelling. These processes play a crucial role in mediating how plants respond to salt stress by modulating gene expression. This often results in heritable changes that can be used as molecular markers. Studies in rice and other cereals have demonstrated a clear association between histone alterations, shifts in DNA methylation patterns, and the expression of salt-responsive genes. Furthermore, epigenetic mechanisms contribute to the development of stress memory, enabling plants to respond more effectively to recurring stressful conditions. Understanding these regulatory pathways offers new opportunities for breeding or engineering salt-tolerant rice varieties, potentially leading to improved crop resilience and productivity under saline conditions.
Full article

Figure 1
Open AccessReview
Convergent Evolution and the Epigenome
by
Sebastian Gaston Alvarado, Annaliese Chang and Maral Tajerian
Epigenomes 2025, 9(4), 45; https://doi.org/10.3390/epigenomes9040045 - 11 Nov 2025
Abstract
Background: Trait convergence or parallelism is widely seen across the animal and plant kingdoms. For example, the evolution of eyes in cephalopods and vertebrate lineages, wings in bats and insects, or shark and dolphin body shapes are examples of convergent evolution. Such traits
[...] Read more.
Background: Trait convergence or parallelism is widely seen across the animal and plant kingdoms. For example, the evolution of eyes in cephalopods and vertebrate lineages, wings in bats and insects, or shark and dolphin body shapes are examples of convergent evolution. Such traits develop as a function of environmental pressures or opportunities that lead to similar outcomes despite the independent origins of underlying tissues, cells, and gene transcriptional patterns. Our current understanding of the molecular processes underlying these phenomena is gene-centric and focuses on how convergence involves the recruitment of novel genes, the recombination of gene products, and the duplication and divergence of genetic substrates. Scope: Despite the independent origins of a given trait, these model organisms still possess some form of epigenetic processes conserved in eukaryotes that mediate gene-by-environment interactions. These traits evolve under similar environmental pressures, so attention should be given to plastic molecular processes that shape gene function along these evolutionary paths. Key Mechanisms: Here, we propose that epigenetic processes such as histone-modifying machinery are essential in mediating the dialog between environment and gene function, leading to trait convergence across disparate lineages. We propose that epigenetic modifications not only mediate gene-by-environment interactions but also bias the distribution of de novo mutations and recombination, thereby channeling evolutionary trajectories toward convergence. An inclusive view of the epigenetic landscape may provide a parsimonious understanding of trait evolution.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures

Figure 1
Open AccessSystematic Review
Exploring the Impact of Nanotherapeutics on Histone H3 and H4 Acetylation Enrichment in Cancer Epigenome: A Systematic Scoping Synthesis
by
Milad Shirvaliloo, Sepideh Khoee, Samideh Khoei, Roghayeh Sheervalilou, Parisa Mohammad Hosseini, Reza Afzalipour and Sakine Shirvalilou
Epigenomes 2025, 9(4), 44; https://doi.org/10.3390/epigenomes9040044 - 7 Nov 2025
Abstract
Background/Objectives: Histone acetylation regulates gene expression and plays a key role in cancer pathophysiology. Nanotherapeutics are known to modulate histone acetylation and influence cancer progression. This systematic scoping review examines the effects of nanotherapeutics on histone acetylation enrichment across multiple cancers. Methods
[...] Read more.
Background/Objectives: Histone acetylation regulates gene expression and plays a key role in cancer pathophysiology. Nanotherapeutics are known to modulate histone acetylation and influence cancer progression. This systematic scoping review examines the effects of nanotherapeutics on histone acetylation enrichment across multiple cancers. Methods: A systematic search of Embase, PubMed/MEDLINE, Scopus, and Web of Science was conducted in accordance with the PRISMA 2020 statement. A total of 13 studies were included. Data were analyzed and visualized in R, and risk of bias was assessed with ToxRTool (OSF Registration: 10.17605/OSF.IO/E643S). Results: Nanotherapeutics were most commonly evaluated against breast (21.4%), prostate (21.4%), pancreatic (14.3%), and bladder (14.3%) cancers. Primary nanomaterials used in the synthesis of nanotherapeutics included poly(lactic-co-glycolic acid) (25.0%), gold (21.4%) and arsenic oxide (21.4%) nanoparticles. Studied histone acetylation marks included H3K9ac, H3K14ac, H3K27ac and H4K16ac. Treatment with nanotherapeutics increased histone H3 and H4 acetylation enrichment, particularly H3K14ac in colorectal and prostate cancers and H4K16ac in ovarian cancer. Conversely, gold-based nanotherapeutics decreased H3K9ac and H3K14ac enrichment in breast cancer. The optimal concentration for most nanotherapeutics was ≤25 µM, with PpIX-FFYSV showing the strongest anticancer effect (viability <25%). Across four preclinical studies (n = 58), treatment with the nanotherapeutics reduced tumor size to less than 50% of control in 64% of animals (95% CI: 21–92%, I2 = 63.8%). Altered histone acetylation was associated with differential expression of CDKN1A, HSPA1, SREBF2 and TGFB. Conclusions: The evidence demonstrates that nanotherapeutics can alter histone acetylation patterns by modulating EP300/CBP, GCN5 and HDAC, preventing cancer progression and invasion.
Full article
(This article belongs to the Special Issue Epigenetic Signatures in Metabolic Health and Cancer)
►▼
Show Figures

Graphical abstract
Open AccessReview
Epigenetic Mechanisms of Plant Adaptation to Cadmium and Heavy Metal Stress
by
Eleonora Greco, Emanuela Talarico, Francesco Guarasci, Marina Camoli, Anna Maria Palermo, Alice Zambelli, Adriana Chiappetta, Fabrizio Araniti and Leonardo Bruno
Epigenomes 2025, 9(4), 43; https://doi.org/10.3390/epigenomes9040043 - 2 Nov 2025
Cited by 3
Abstract
►▼
Show Figures
Heavy metal and metalloid stress, particularly from toxic elements like cadmium (Cd), poses a growing threat to plant ecosystems, crop productivity, and global food security. Elevated concentrations of these contaminants can trigger cytotoxic and genotoxic effects in plants, severely impairing growth, development, and
[...] Read more.
Heavy metal and metalloid stress, particularly from toxic elements like cadmium (Cd), poses a growing threat to plant ecosystems, crop productivity, and global food security. Elevated concentrations of these contaminants can trigger cytotoxic and genotoxic effects in plants, severely impairing growth, development, and reproduction. In recent years, epigenetic mechanisms have emerged as crucial regulators of plant responses to heavy metal stress, offering novel insights and strategies for enhancing plant resilience in contaminated environments. This review synthesises current advances in the field of plant epigenetics, focusing on key modifications such as DNA methylation, histone acetylation and remodelling, chromatin dynamics, and small RNA-mediated regulation. These processes not only influence gene expression under metal-induced stress but also hold promise for long-term adaptation through transgenerational epigenetic memory. Recent developments in high-throughput sequencing and functional genomics have accelerated the identification of epigenetic markers associated with stress tolerance, enabling the integration of these markers into breeding programs and targeted epigenome editing strategies. Special attention is given to cadmium stress responses, where specific epigenetic traits have been linked to enhanced tolerance. As plant epigenomic research progresses, its application in sustainable agriculture becomes increasingly evident offering environmentally friendly solutions to mitigate the impact of heavy metal pollution. This review provides a foundation for future research aimed at leveraging epigenetic tools to engineer crops capable of thriving under metal stress, thereby contributing to resilient agricultural systems and sustainable food production.
Full article

Figure 1

Journal Menu
► ▼ Journal Menu-
- Epigenomes Home
- Aims & Scope
- Editorial Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Topical Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Brain Sciences, CIMB, Epigenomes, Genes, IJMS, DNA
Genetics and Epigenetics of Substance Use Disorders
Topic Editors: Aleksandra Suchanecka, Anna Maria Grzywacz, Kszysztof ChmielowiecDeadline: 14 April 2027

Special Issues
Special Issue in
Epigenomes
Epigenetic Signatures in Metabolic Health and Cancer
Guest Editor: Aparna DuggiralaDeadline: 30 April 2026
Special Issue in
Epigenomes
DNA Methylation Markers in Health and Disease
Guest Editor: Osman El‐MaarriDeadline: 31 May 2026
Special Issue in
Epigenomes
Single-Cell and Spatial Epigenomics: Methods and Applications
Guest Editor: Sankarasubramanian JagadesanDeadline: 20 June 2026
Special Issue in
Epigenomes
Epigenetics Meets Immunology: Mechanisms, Crosstalk, and Therapeutic Implications
Guest Editor: Yalong WangDeadline: 31 July 2026
Topical Collections
Topical Collection in
Epigenomes
Epigenetic Mechanisms in Diabetes Research
Collection Editor: Assam El-Osta
Topical Collection in
Epigenomes
Feature Papers in Epigenomes
Collection Editors: Ivana De la Serna, Che-Kun James Shen








