Amino Acid Metabolism in Leukocytes Showing In Vitro IgG Memory from SARS-CoV2-Infected Patients
Abstract
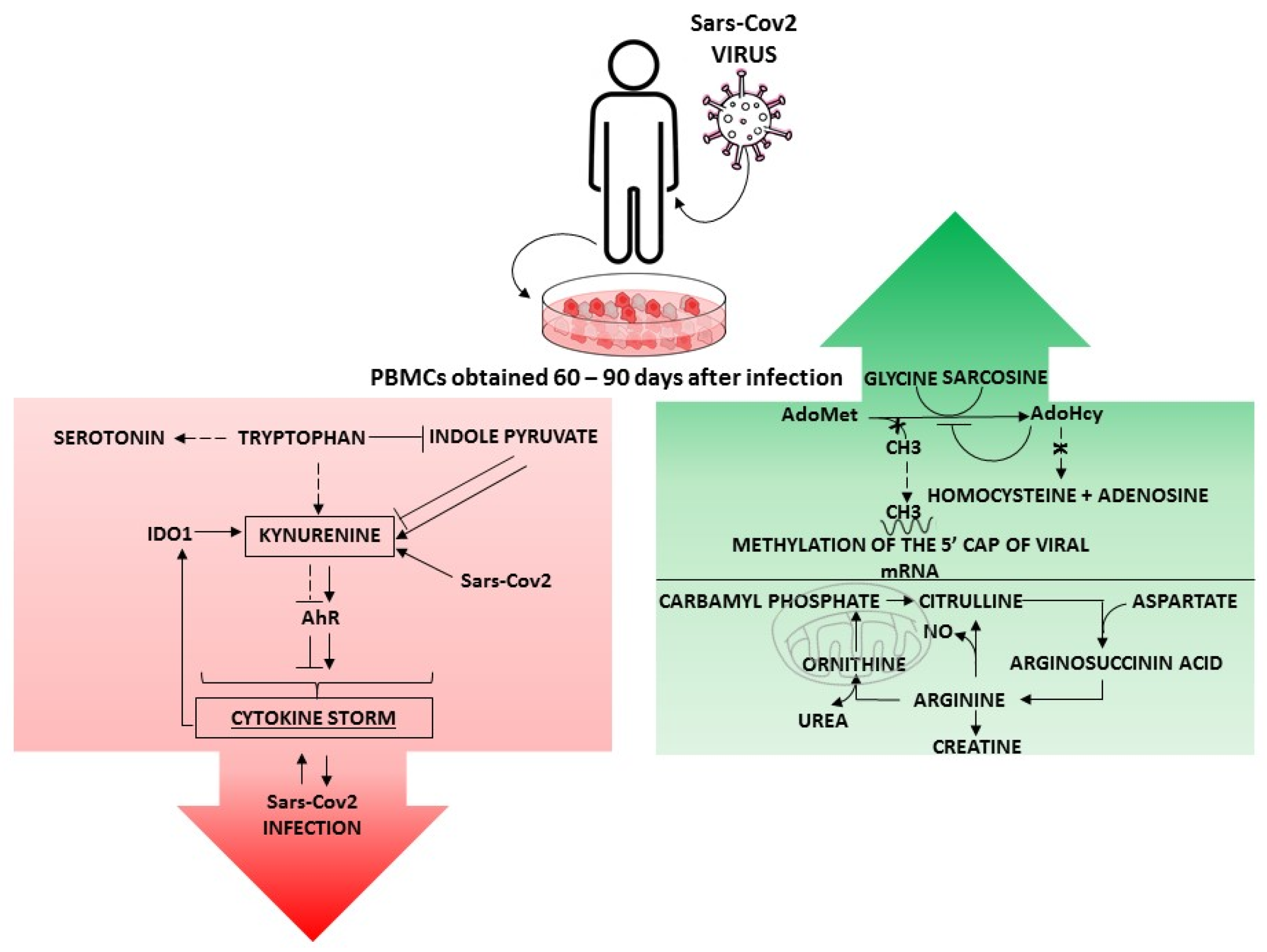
:1. Metabolic Rewiring in SARS-CoV2-Infected Patients
2. Amino Acid Metabolism
2.1. Methionine Cycle
2.2. Arginine Metabolism
2.3. Tryptophan Metabolism
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Singh, A.; Shaikh, A.; Singh, R.; Singh, A.K. COVID-19: From bench to bed side. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 277–281. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Coronavirus Disease (COVID-2019) Situation Report-140. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 13 April 2023).
- D’amico, F.; Baumgart, D.C.; Danese, S.; Peyrin-Biroulet, L. Diarrhea during COVID-19 infection: Pathogenesis, epidemiology, prevention, and management. Clin. Gastroenterol. Hepatol. 2020, 18, 1663–1672. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Walker, A.; Pottinger, G.; Scott, A.; Hopkins, C. Anosmia and loss of smell in the era of COVID-19. BMJ 2020, 370, m2808. [Google Scholar] [CrossRef] [PubMed]
- Soler, Z.M.; Patel, Z.M.; Turner, J.H.; Holbrook, E.H. A primer on viral-associated olfactory loss in the era of COVID-19. Int. Forum Allergy Rhinol. 2020, 10, 814–820. [Google Scholar] [CrossRef]
- Schwab, J.; Fjaeldstad, A.W. Recovery rates and parosmia in olfactory loss during the COVID-19 era. Dan. Med. J. 2022, 69, A04220271. [Google Scholar]
- Moein, S.T.; Hashemian, S.M.; Mansourafshar, B.; Khorram-Tousi, A.; Tabarsi, P.; Doty, R.L. Smell dysfunction: A biomarker for COVID-19. Int. Forum Allergy Rhinol. 2020, 10, 944–950. [Google Scholar] [CrossRef]
- Ahn, D.-G.; Shin, H.-J.; Kim, M.-H.; Lee, S.; Kim, H.-S.; Myoung, J.; Kim, B.-T.; Kim, S.-J. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19). J. Microbiol. Biotechnol. 2020, 30, 313–324. [Google Scholar] [CrossRef]
- Thye, A.Y.; Law, J.W.; Pusparajah, P.; Letchumanan, V.; Chan, K.G.; Lee, L.H. Emerging SARS-CoV2 Variants of Concern (VOCs): An Impending Global Crisis. Biomedicines 2021, 9, 1303. [Google Scholar] [CrossRef]
- Kläser, K.; Molteni, E.; Graham, M.; Canas, L.S.; Österdahl, M.F.; Antonelli, M.; Chen, L.; Deng, J.; Murray, B.; Kerfoot, E.; et al. COVID-19 due to the B.1.617.2 (Delta) variant compared to B.1.1.7 (Alpha) variant of SARS-CoV2: A prospective observational cohort study. Sci. Rep. 2022, 12, 10904. [Google Scholar] [CrossRef]
- Ren, S.Y.; Wang, W.B.; Gao, R.D.; Zhou, A.M. Omicron variant (B.1.1.529) of SARS-CoV2: Mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases 2022, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.H.; Faraji, F.; Prajapati, D.P.; Boone, C.E.; DeConde, A.S. Association of chemosensory dysfunction and COVID-19 in patients presenting with influenza-like symptoms. Int. Forum Allergy Rhinol. 2020, 10, 806–813. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Chen, Y.; Qin, Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020, 92, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.-J.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Jeon, S.; Shin, H.Y.; Kim, M.J.; Seong, Y.M.; Lee, W.J.; Choe, K.W.; Kang, Y.M.; Lee, B.; Park, S.J. Case of the index patient who caused tertiary transmission of Coronavirus disease 2019 in Korea: The application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCR. Korean Med. Sci. 2020, 35, e79. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Yu, X.; Du, X.; Li, Q.; Li, X.; Qin, T.; Wang, M.; Jiang, M.; Li, J.; Li, W.; et al. Epidemiological and clinical characteristics of 26 asymptomatic SARS-CoV-2 carriers. J. Infect. Dis. 2020, 221, 1940–1947. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Ye, F.; Cheng, M.L.; Feng, Y.; Deng, Y.Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X.; et al. Detection of SARS-CoV-2-specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020, 52, 971–977.e3. [Google Scholar] [CrossRef]
- Zarletti, G.; Tiberi, M.; De Molfetta, V.; Bossù, M.; Toppi, E.; Bossù, P.; Scapigliati, G. A Cell-Based ELISA to Improve the Serological Analysis of Anti-SARS-CoV-2 IgG. Viruses 2020, 12, 1274. [Google Scholar] [CrossRef]
- Fanelli, G.; Gevi, F.; Zarletti, G.; Tiberi, M.; De Molfetta, V.; Scapigliati, G.; Timperio, A.M. An Altered Metabolism in Leukocytes Showing in vitro igG Memory from SARS-CoV-2-Infected Patients. Front. Mol. Biosci. 2022, 9, 894207. [Google Scholar] [CrossRef]
- Mahmudpour, M.; Roozbeh, J.; Keshavarz, M.; Farrokhi, S.; Nabipour, I. COVID-19 cytokine storm: The anger of inflammation. Cytokine 2020, 133, 155151. [Google Scholar] [CrossRef]
- Federica, G.; Giuseppina, F.; Veronica, L.; Gianpaolo, Z.; Massimo, T.; Veronica, D.M.; Giuseppe, S.; Maria, T.A. An untargeted metabolomic approach to investigate antiviral defence mechanisms in memory leukocytes secreting anti-SARS-CoV-2 IgG in vitro. Sci. Rep. 2023, 13, 629. [Google Scholar] [CrossRef]
- Liptak, P.; Baranovicova, E.; Rosolanka, R.; Simekova, K.; Bobcakova, A.; Vysehradsky, R.; Duricek, M.; Dankova, Z.; Kapinova, A.; Dvorska, D.; et al. Persistence of Metabolomic Changes in Patients during Post-COVID Phase: A Prospective, Observational Study. Metabolites 2022, 12, 641. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Luo, P.; Xu, J.; Yang, L.; Ma, P.; Tan, X.; Chen, Q.; Zhou, M.; Song, S.; Xia, H.; et al. Plasma Metabolomic Profiles in Recovered COVID-19 Patients without Previous Underlying Diseases 3 Months after Discharge. J. Inflamm. Res. 2021, 14, 4485–4501. [Google Scholar] [CrossRef] [PubMed]
- Danlos, F.-X.; Grajeda-Iglesias, C.; Durand, S.; Sauvat, A.; Roumier, M.; Cantin, D.; Colomba, E.; Rohmer, J.; Pommeret, F.; Baciarello, G.; et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 2022, 12, 258. [Google Scholar] [CrossRef]
- Bruzzone, C.; Conde, R.; Embade, N.; Mato, J.M.; Millet, O. Metabolomics as a powerful tool for diagnostic, pronostic and drug intervention analysis in COVID-19. Front. Mol. Biosci. 2023, 10, 1111482. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Caterino, M.; Fedele, R.; Cevenini, A.; Pontillo, M.; Barra, L.; Ruoppolo, M. COVIDomics: The Proteomic and Metabolomic Signatures of COVID-19. Int. J. Mol. Sci. 2022, 23, 2414. [Google Scholar] [CrossRef] [PubMed]
- Gowda, G.N.; Zhang, S.; Gu, H.; Asiago, V.; Shanaiah, N.; Raftery, D. Metabolomics-based methods for early disease diagnostics. Expert. Rev. Mol. Diagn. 2008, 8, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Borba, M.G.; Val, F.F.; Sampaio, V.S.; Alexandre, M.A.; Melo, G.C.; Brito, M.; Mourão, M.P.; Brito-Sousa, J.D.; Baía-da-Silva, D.; Guerra, M.V.; et al. Effect of High vs. Low Doses of Chloroquine Diphosphate as Adjunctive Therapy for Patients Hospitalized with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection: A Randomized Clinical Trial. JAMA Netw. Open 2020, 3, e208857. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Xu, G.; Xie, Y.; Wang, X.; Wu, J.; Chen, H. Plasma metabolomic characterization of SARS-CoV-2 Omicron infection. Cell Death Dis. 2023, 14, 276. [Google Scholar] [CrossRef]
- Shen, T.; Wang, T. Metabolic Reprogramming in COVID-19. Int. J. Mol. Sci. 2021, 22, 11475. [Google Scholar] [CrossRef]
- Maltais-Payette, I.; Lajeunesse-Trempe, F.; Pibarot, P.; Biertho, L.; Tchernof, A. Association between Circulating Amino Acids and COVID-19 Severity. Metabolites 2023, 13, 201. [Google Scholar] [CrossRef] [PubMed]
- Maeda, R.; Seki, N.; Uwamino, Y.; Wakui, M.; Nakagama, Y.; Kido, Y.; Sasai, M.; Taira, S.; Toriu, N.; Yamamoto, M.; et al. Amino acid catabolite markers for early prognostication of pneumonia in patients with COVID-19. Nat. Commun. 2023, 14, 8469. [Google Scholar] [CrossRef] [PubMed]
- Ansone, L.; Briviba, M.; Silamikelis, I.; Terentjeva, A.; Perkons, I.; Birzniece, L.; Rovite, V.; Rozentale, B.; Viksna, L.; Kolesova, O.; et al. Amino Acid Metabolism is Significantly Altered at the Time of Admission in Hospital for Severe COVID-19 Patients: Findings from Longitudinal Targeted Metabolomics Analysis. Microbiol. Spectr. 2021, 9, e0033821. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yin, Y.-L.; Li, D.; Kim, S.W.; Wu, G. Amino acids and immune function. Br. J. Nutr. 2007, 98, 237–252. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gómez, L.E.; Ibarra-González, I.; Fernández-Lainez, C.; Tusie, T.; Moreno-Macías, H.; Martinez-Armenta, C.; Jimenez-Gutierrez, G.E.; Vázquez-Cárdenas, P.; Vidal-Vázquez, P.; Ramírez-Hinojosa, J.P.; et al. Metabolic Reprogramming in SARS-CoV-2 Infection Impacts the Outcome of COVID-19 Patients. Front. Immunol. 2022, 13, 936106. [Google Scholar] [CrossRef] [PubMed]
- Páez-Franco, J.C.; Torres-Ruiz, J.; Sosa-Hernández, V.A.; Cervantes-Díaz, R.; Romero-Ramírez, S.; Pérez-Fragoso, A.; Meza-Sánchez, D.E.; Germán-Acacio, J.M.; Maravillas-Montero, J.L.; Mejía-Domínguez, N.R.; et al. Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients. Sci. Rep. 2021, 11, 6350. [Google Scholar] [CrossRef] [PubMed]
- Halliley, J.L.; Kyu, S.; Kobie, J.J.; Walsh, E.E.; Falsey, A.R.; Randall, T.D.; Treanor, J.; Feng, C.; Sanz, I.; Lee, F.E.-H. Peak frequencies of circulating human influenza-specific antibody secreting cells correlate with serum antibody response after immunization. Vaccine 2010, 28, 3582–3587. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Buisman, A.; de Rond, C.; Öztürk, K.; Hulscher, H.T.; van Binnendijk, R. Long-term presence of memory B-cells specific for different vaccine components. Vaccine 2009, 28, 179–186. [Google Scholar] [CrossRef]
- Beavis, K.G.; Matushek, S.M.; Abeleda, A.P.; Bethel, C.; Hunt, C.; Gillen, S.; Moran, A.; Tesic, V. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA Assay for detection of IgA and IgG antibodies. J. Clin. Virol. 2020, 129, 104468. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Rhea, E.M.; Logsdon, A.F.; Hansen, K.M.; Williams, L.M.; Reed, M.J.; Baumann, K.K.; Holden, S.J.; Raber, J.; Banks, W.A.; Erickson, M.A. The S1 protein of SARS-CoV-2 crosses the blood–brain barrier in mice. Nat. Neurosci. 2021, 24, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Laing, A.G.; Lorenc, A.; del Barrio, I.D.M.; Das, A.; Fish, M.; Monin, L.; Muñoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020, 26, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.-N.; Jiang, Y.-F.; Ru, J.-N.; Lu, J.-H.; Ding, B.; Wu, J. Amino acid metabolism in health and disease. Signal Transduct. Target. Ther. 2023, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, S.; Lee, J.; Jang, J.; Jung, J.; Koh, J.-H.; Choi, C.S.; Wolfe, R.R.; Kim, I.-Y. Essential Amino Acid-Enriched Diet Alleviates Dexamethasone-Induced Loss of Muscle Mass and Function through Stimulation of Myofibrillar Protein Synthesis and Improves Glucose Metabolism in Mice. Metabolites 2022, 12, 84. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, A.; Bayraktar, N.; Bayraktar, M.; Ibrahim, B.; Bozok, T.; Resat, C. Evaluation of amino acid profile in serum of patients with COVID-19 for providing a new treatment strategy. J. Med. Biochem. 2022, 41, 526–533. [Google Scholar] [CrossRef]
- Schiuma, G.; Beltrami, S.; Bortolotti, D.; Rizzo, R.; Rizzo, S. Innate Immune Response in SARS-CoV-2 Infection. Microorganisms 2022, 10, 501. [Google Scholar] [CrossRef]
- Atila, A.; Alay, H.; Yaman, M.E.; Akman, T.C.; Cadirci, E.; Bayrak, B.; Celik, S.; Atila, N.E.; Yaganoglu, A.M.; Kadioglu, Y.; et al. The serum amino acid profile in COVID-19. Amino Acids 2021, 53, 1569–1588. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Wu, J.; Zhao, M.; Li, C.; Zhang, Y.; Wang, D.W. The SARS-CoV-2 induced targeted amino acid profiling in patients at hospitalized and convalescent stage. Biosci. Rep. 2021, 41, BSR20204201. [Google Scholar] [CrossRef] [PubMed]
- Benavides, M.A. l-Methionine may modulate the assembly of SARS-CoV-2 by interfering with the mechanism of RNA polymerase. Med. Hypotheses 2022, 161, 110798. [Google Scholar] [CrossRef] [PubMed]
- Perła-Kaján, J.; Jakubowski, H. COVID-19 and One-Carbon Metabolism. Int. J. Mol. Sci. 2022, 23, 4181. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Ren, D.; Jeon, B.; Liu, H.-W. S-Adenosylmethionine: More than just a methyl donor. Nat. Prod. Rep. 2023, 40, 1521–1549. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C. S-Adenosylmethionine. Int. J. Biochem. Cell Biol. 2000, 32, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Y.; Wu, A.; Xu, S.; Pan, R.; Zeng, C.; Jin, X.; Ge, X.; Shi, Z.; Ahola, T.; et al. Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J. Virol. 2015, 89, 8416–8427. [Google Scholar] [CrossRef] [PubMed]
- Aouadi, W.; Blanjoie, A.; Vasseur, J.-J.; Debart, F.; Canard, B.; Decroly, E. Binding of the methyl donor S-adenosyl-L-methionine to Middle East respiratory syndrome coronavirus 2′-O-methyltransferase nsp 16 promotes recruitment of the allosteric activator nsp10. J. Virol. 2017, 91, e02217-16. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M.; Han, Q. Oral Methioninase for COVID-19 Methionine-restriction Therapy. In Vivo 2020, 34, 1593–1596. [Google Scholar] [CrossRef]
- Yan, L.; Yang, Y.; Li, M.; Zhang, Y.; Zheng, L.; Ge, J.; Huang, Y.C.; Liu, Z.; Wang, T.; Gao, S.; et al. Coupling of N7-methyltransferase and 3′-5′ exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell 2021, 184, 3474–3485.e11. [Google Scholar] [CrossRef]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef]
- Byszewska, M.; Śmietański, M.; Purta, E.; Bujnicki, J.M. RNA Methyltransferases Involved in 5′ Cap Biosynthesis. RNA Biol. 2014, 11, 1597–1607. [Google Scholar] [CrossRef] [PubMed]
- Kryukov, E.V.; Ivanov, A.V.; Karpov, V.O.; Aleksandrin, V.V.; Dygai, A.M.; Kruglova, M.P.; Kostiuchenko, G.I.; Kazakov, S.P.; Kubatiev, A.A. Plasma S-Adenosylmethionine Is Associated with Lung Injury in COVID-19. Dis. Markers 2021, 2021, 7686374. [Google Scholar] [CrossRef] [PubMed]
- Zulet, M.I.; Fontes, L.P.; Blanco, T.A.; Bescos, F.L.; Iriarte, M.M. Epigenetic changes in neurology: DNA methylation in multiple sclerosis. Modificaciones epigenéticas en neurología: Alteraciones en la metilación del ADN en la esclerosis múltiple. Neurologia 2017, 32, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Stollar, V.; Mensah, V.; Adams, S.; Li, M.-L. Evolution of sindbis virus with a low-methionine-resistant phenotype is dependent both on a pre-existing mutation and on the methionine concentration in the medium. PLoS ONE 2013, 8, e60504. [Google Scholar] [CrossRef] [PubMed]
- Villalón, M.D.G.; Gil-Fernández, C.; De Clercq, E. The Activity of Several S-Adenosylhomocysteine Hydrolase Inhibitors against African Swine Fever Virus Replication in Vero Cells. Antivir. Res. 1993, 20, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.; Driscoll, J.; Huggins, J.W. Treatment of Lethal Ebola Virus Infection in Mice with a Single Dose of an S-Adenosyl-L-Homocysteine Hydrolase Inhibitor. Antivir. Res. 2000, 45, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Adebayo, A.; Varzideh, F.; Wilson, S.; Gambardella, J.; Eacobacci, M.; Jankauskas, S.S.; Donkor, K.; Kansakar, U.; Trimarco, V.; Mone, P.; et al. l-Arginine and COVID-19: An Update. Nutrients 2021, 13, 3951. [Google Scholar] [CrossRef] [PubMed]
- Derakhshani, A.; Hemmat, N.; Asadzadeh, Z.; Ghaseminia, M.; Shadbad, M.A.; Jadideslam, G.; Silvestris, N.; Racanelli, V.; Baradaran, B. Arginase 1 (Arg1) as an Up-Regulated Gene in COVID-19 Patients: A Promising Marker in COVID-19 Immunopathy. J. Clin. Med. 2021, 10, 1051. [Google Scholar] [CrossRef]
- Izzo, R.; Trimarco, V.; Mone, P.; Aloè, T.; Marzani, M.C.; Diana, A.; Fazio, G.; Mallardo, M.; Maniscalco, M.; Marazzi, G.; et al. Combining L-Arginine with vitamin C improves long-COVID symptoms: The LINCOLN Survey. Pharmacol. Res. 2022, 183, 106360. [Google Scholar] [CrossRef]
- Paneroni, M.; Pasini, E.; Vitacca, M.; Scalvini, S.; Comini, L.; Pedrinolla, A.; Venturelli, M. Altered Vascular Endothelium-Dependent Responsiveness in Frail Elderly Patients Recovering from COVID-19 Pneumonia: Preliminary Evidence. J. Clin. Med. 2021, 10, 2558. [Google Scholar] [CrossRef]
- Wu, G.; Morris, S.M., Jr. Arginine metabolism: Nitric oxide and beyond. Biochem. J. 1998, 336, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Watford, M. The urea cycle: Teaching intermediary metabolism in a physiological setting. Biochem. Mol. Biol. Educ. 2003, 31, 289–297. [Google Scholar] [CrossRef]
- Koga, T.; Zhang, W.Y.; Gotoh, T.; Oyadomari, S.; Tanihara, H.; Mori, M. Induction of citrulline–nitric oxide (NO) cycle enzymes and NO production in immunostimulated rat RPE-J cells. Exp. Eye Res. 2003, 76, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Bazer, F.W.; Davis, T.A.; Kim, S.W.; Li, P.; Rhoads, J.M.; Satterfield, M.C.; Smith, S.B.; Spencer, T.E.; Yin, Y. Arginine metabolism and nutrition in growth, health and disease. Amino Acids 2009, 37, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C. Nitric oxide as a secretory product of mammalian cells. FASEB J. 1992, 6, 3051–3064. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, C. Nitric oxide and the immune response. Nat. Immunol. 2001, 2, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Marshall, H.E.; Merchant, K.; Stamler, J.S. Nitrosation and oxidation in the regulation of gene expression. FASEB J. 2000, 14, 1889–1900. [Google Scholar] [CrossRef]
- Bogdan, C. The function of nitric oxide in the immune system. Handbook of Experimental Pharmacology. In Nitric Oxide; Mayer, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2000; pp. 443–492. [Google Scholar]
- Song, P.; Li, W.; Xie, J.; Hou, Y.; You, C. Cytokine storm induced by SARS-CoV-2. Clin. Chim. Acta 2020, 509, 280–287. [Google Scholar] [CrossRef]
- Shenoy, S. Coronavirus (COVID-19) sepsis: Revisiting mitochondrial dysfunction in pathogenesis, aging, inflammation, and mortality. Inflamm. Res. 2020, 69, 1077–1085. [Google Scholar] [CrossRef]
- Nambiar, V.; Sudevan, R.; Ajai, A.; Chattu, V.K. Growing burden of stroke, recent advancements in management and global commitments: The way forward. J. Pharm. Pract. Community Med. 2018, 4, 191–192. [Google Scholar] [CrossRef]
- Uehara, E.U.; Shida Bde, S.; de Brito, C.A. Role of nitric oxide in immune responses against viruses: Beyond microbicidal activity. Inflamm. Res. 2015, 64, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Åkerström, S.; Mousavi-Jazi, M.; Klingström, J.; Leijon, M.; Lundkvist, A.; Mirazimi, A. Nitric oxide inhibits the replication cycle of severe acute respiratory syndrome coronavirus. J. Virol. 2005, 79, 1966–1969. [Google Scholar] [CrossRef] [PubMed]
- Akaberi, D.; Krambrich, J.; Ling, J.; Luni, C.; Hedenstierna, G.; Järhult, J.D.; Lennerstrand, J.; Lundkvist, Å. Mitigation of the replication of SARS-CoV-2 by nitric oxide in vitro. Redox Biol. 2020, 37, 101734. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; Chathambath, A.; Al-Sehemi, A.G.; Pannipara, M.; Unnikrishnan, M.K.; Aleya, L.; Raghavan, R.P.; Mathew, B. Critical role of nitric oxide in impeding COVID-19 transmission and prevention: A promising possibility. Environ. Sci. Pollut. Res. 2022, 29, 38657–38672. [Google Scholar] [CrossRef] [PubMed]
- Rees, C.A.; Rostad, C.A.; Mantus, G.; Anderson, E.J.; Chahroudi, A.; Jaggi, P.; Wrammert, J.; Ochoa, J.B.; Ochoa, A.; Basu, R.K.; et al. Altered amino acid profile in patients with SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2021, 118, e210170811. [Google Scholar] [CrossRef] [PubMed]
- Sacchi, A.; Grassi, G.; Notari, S.; Gili, S.; Bordoni, V.; Tartaglia, E.; Casetti, R.; Cimini, E.; Mariotti, D.; Garotto, G.; et al. Expansion of Myeloid Derived Suppressor Cells Contributes to Platelet Activation by L-Arginine Deprivation during SARS-CoV-2 Infection. Cell 2021, 10, 2111. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, J.B.; Bernard, A.C.; O’brien, W.E.; Griffen, M.M.; Maley, M.E.; Rockich, A.K.; Tsuei, B.J.; Boulanger, B.R.; Kearney, P.A.; Morris, S.M. Arginase I expression and activity in human mononuclear cells after injury. Ann. Surg. 2001, 233, 393–399. [Google Scholar] [CrossRef]
- Takeshita, H.; Yamamoto, K. Tryptophan Metabolism and COVID-19-Induced Skeletal Muscle Damage: Is ACE2 a Key Regulator? Front. Nutr. 2022, 9, 868845. [Google Scholar] [CrossRef]
- Gardinassi, L.G.; Souza, C.O.S.; Sales-Campos, H.; Fonseca, S.G. Immune and Metabolic Signatures of COVID-19 Revealed by Transcriptomics Data Reuse. Front. Immunol. 2020, 11, 1636. [Google Scholar] [CrossRef]
- Achtyes, E.; Keaton, S.A.; Smart, L.; Burmeister, A.R.; Heilman, P.L.; Krzyzanowski, S.; Nagalla, M.; Guillemin, G.J.; Galvis, M.L.E.; Lim, C.K.; et al. Inflammation and kynurenine pathway dysregulation in post-partum women with severe and suicidal depression. Brain Behav. Immun. 2020, 83, 239–247. [Google Scholar] [CrossRef]
- Platten, M.; Nollen, E.A.A.; Röhrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef] [PubMed]
- Modoux, M.; Rolhion, N.; Mani, S.; Sokol, H. Tryptophan Metabolism as a Pharmacological Target. Trends Pharmacol. Sci. 2021, 42, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Guarnieri, T. Hypothesis: Emerging Roles for Aryl Hydrocarbon Receptor in Orchestrating CoV-2-Related Inflammation. Cells 2022, 11, 648. [Google Scholar] [CrossRef] [PubMed]
- Pallotta, M.T.; Orabona, C.; Volpi, C.; Vacca, C.; Belladonna, M.L.; Bianchi, R.; Servillo, G.; Brunacci, C.; Calvitti, M.; Bicciato, S.; et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat. Immunol. 2011, 12, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Cervenka, I.; Agudelo, L.Z.; Ruas, J.L. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science 2017, 357, eaaf9794. [Google Scholar] [CrossRef] [PubMed]
- Lionetto, L.; Ulivieri, M.; Capi, M.; De Bernardini, D.; Fazio, F.; Petrucca, A.; Pomes, L.M.; De Luca, O.; Gentile, G.; Casolla, B.; et al. Increased kynurenine-to-tryptophan ratio in the serum of patients infected with SARS-CoV2: An observational cohort study. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166042. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Nie, M.; Pang, H.; Wang, B.; Hu, J.; Meng, X.; Li, K.; Ran, X.; Long, Q.; Deng, H.; et al. Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 2021, 12, 1618. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.; Anderson, G. False dogmas in schizophrenia research: Toward the reifcation of pathway phenotypes and pathway classes. Front. Psychiatry 2021, 12, 663985. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, J.; Yang, Y.; Ma, H.; Li, Z.; Zhang, J.; Cheng, J.; Zhang, X.; Zhao, Y.; Xia, Z.; et al. The role of interleukin-6 in monitoring severe case of coronavirus disease 2019. EMBO Mol. Med. 2020, 12, e12421. [Google Scholar] [CrossRef]
- Dehhaghi, M.; Kazemi Shariat Panahi, H.; Guillemin, G.J. Microorganisms, tryptophan metabolism, and kynurenine pathway: A complex interconnected loop influencing human health status. Int. J. Tryptophan Res. 2019, 12, 117864691985299. [Google Scholar] [CrossRef]
- Garcez, M.L.; Jacobs, K.R.; Guillemin, G.J. Microbiota alterations in alzheimer’s disease: Involvement of the kynurenine pathway and inflammation. Neurotox. Res. 2019, 36, 424–436. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanelli, G.; Lelli, V.; Rinalducci, S.; Timperio, A.M. Amino Acid Metabolism in Leukocytes Showing In Vitro IgG Memory from SARS-CoV2-Infected Patients. Diseases 2024, 12, 43. https://doi.org/10.3390/diseases12030043
Fanelli G, Lelli V, Rinalducci S, Timperio AM. Amino Acid Metabolism in Leukocytes Showing In Vitro IgG Memory from SARS-CoV2-Infected Patients. Diseases. 2024; 12(3):43. https://doi.org/10.3390/diseases12030043
Chicago/Turabian StyleFanelli, Giuseppina, Veronica Lelli, Sara Rinalducci, and Anna Maria Timperio. 2024. "Amino Acid Metabolism in Leukocytes Showing In Vitro IgG Memory from SARS-CoV2-Infected Patients" Diseases 12, no. 3: 43. https://doi.org/10.3390/diseases12030043
APA StyleFanelli, G., Lelli, V., Rinalducci, S., & Timperio, A. M. (2024). Amino Acid Metabolism in Leukocytes Showing In Vitro IgG Memory from SARS-CoV2-Infected Patients. Diseases, 12(3), 43. https://doi.org/10.3390/diseases12030043







