Nanobiosensors for the Detection of Novel Coronavirus 2019-nCoV and Other Pandemic/Epidemic Respiratory Viruses: A Review
Abstract
:1. Introduction
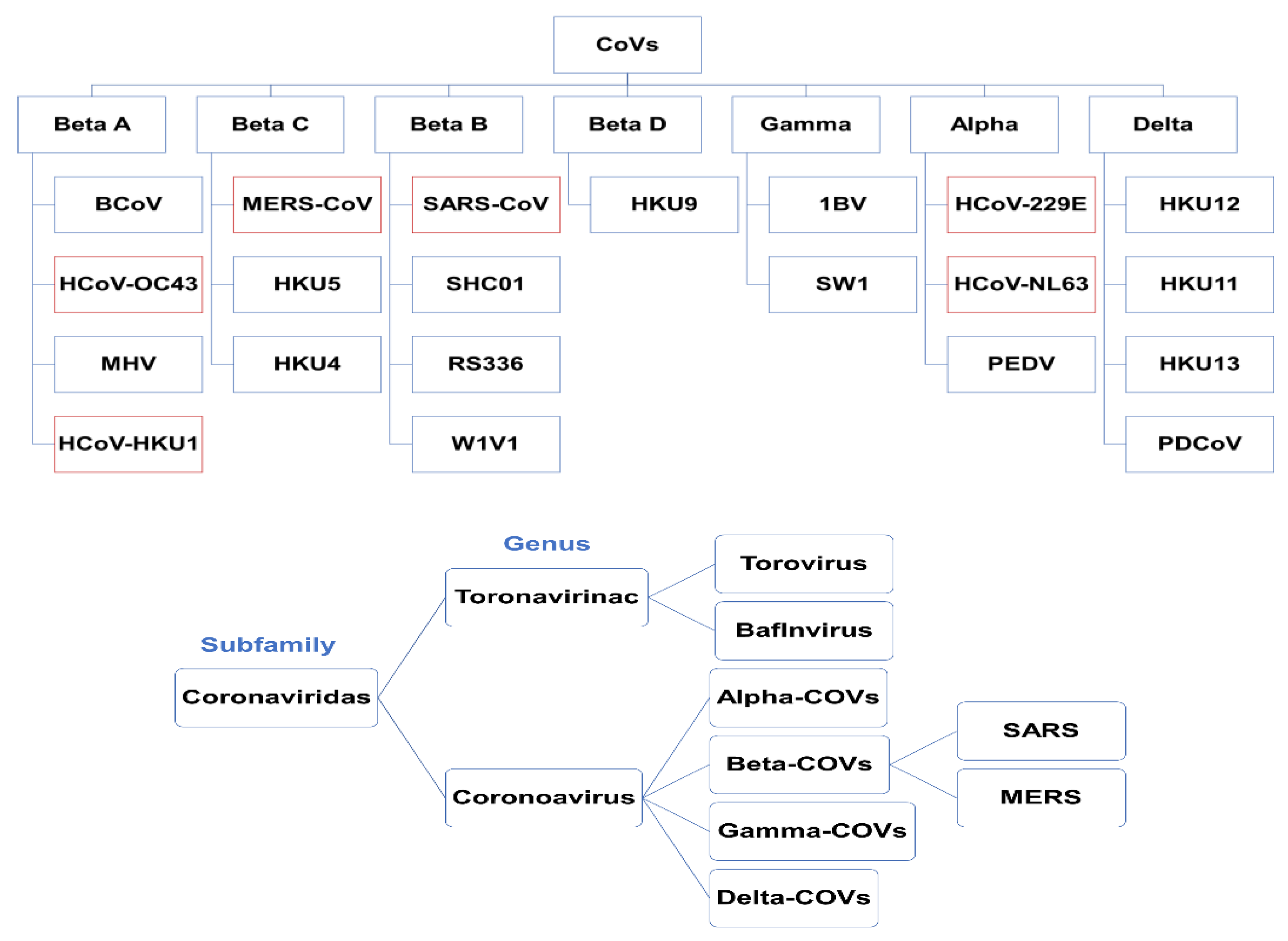
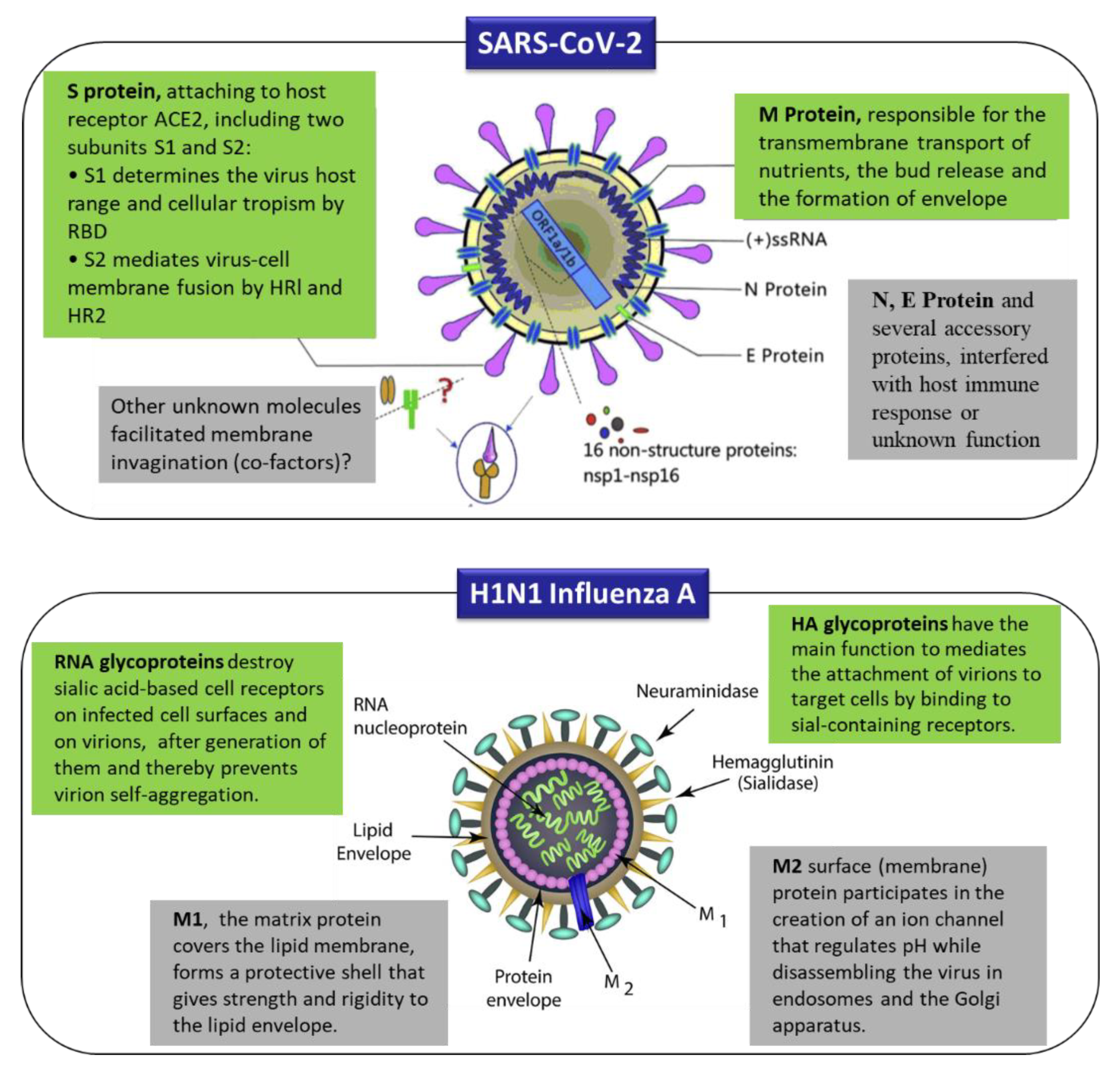
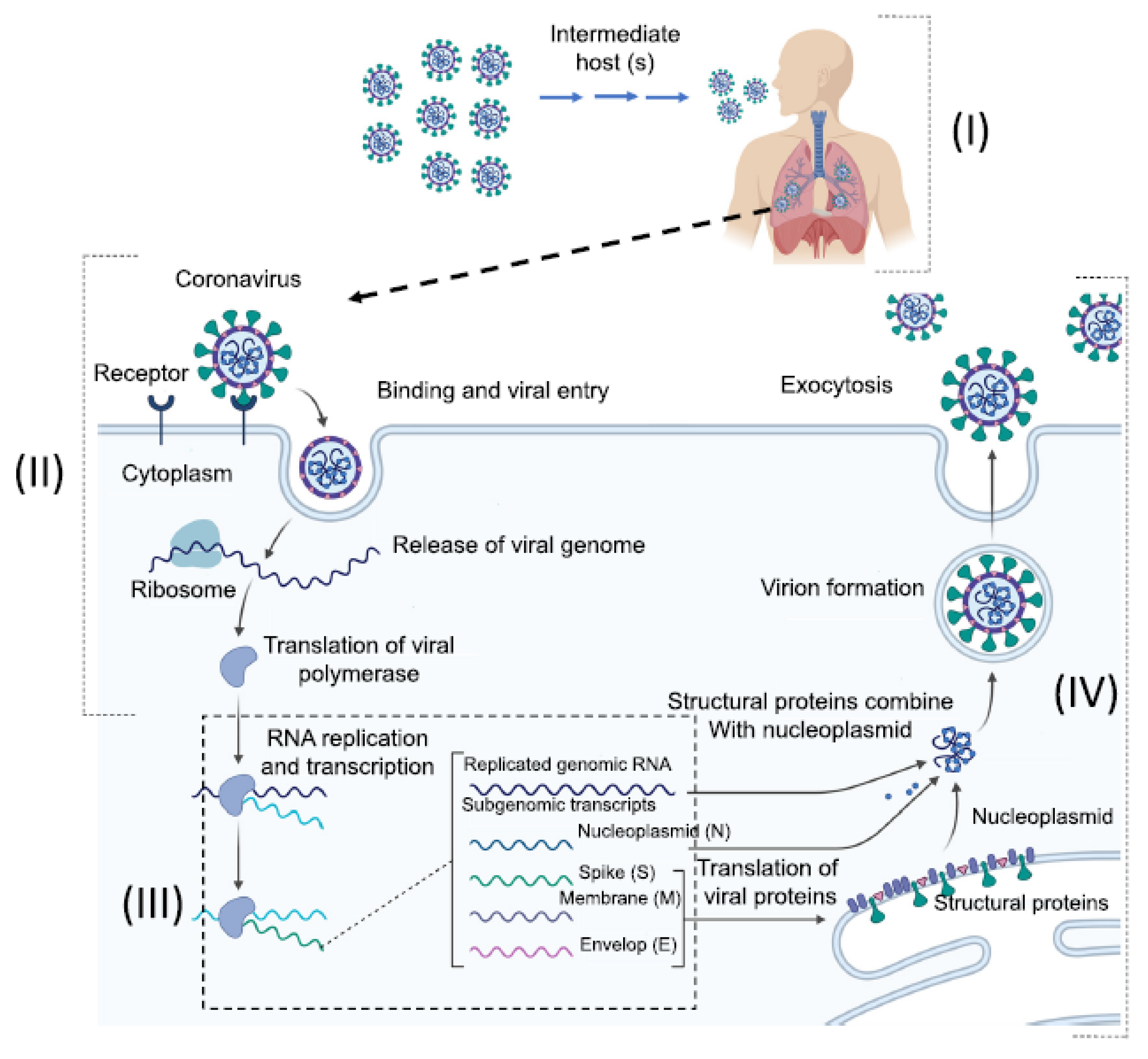
2. Origins, Classifications, and Structures of Human Coronaviruses
3. Significance of Biological Receptors
4. Nanobiosensors for the Detection of Human Coronavirus (2019-nCoV, SARS/MERS-CoV) and Influenza Viruses
4.1. Electrochemical Nanobiosensors
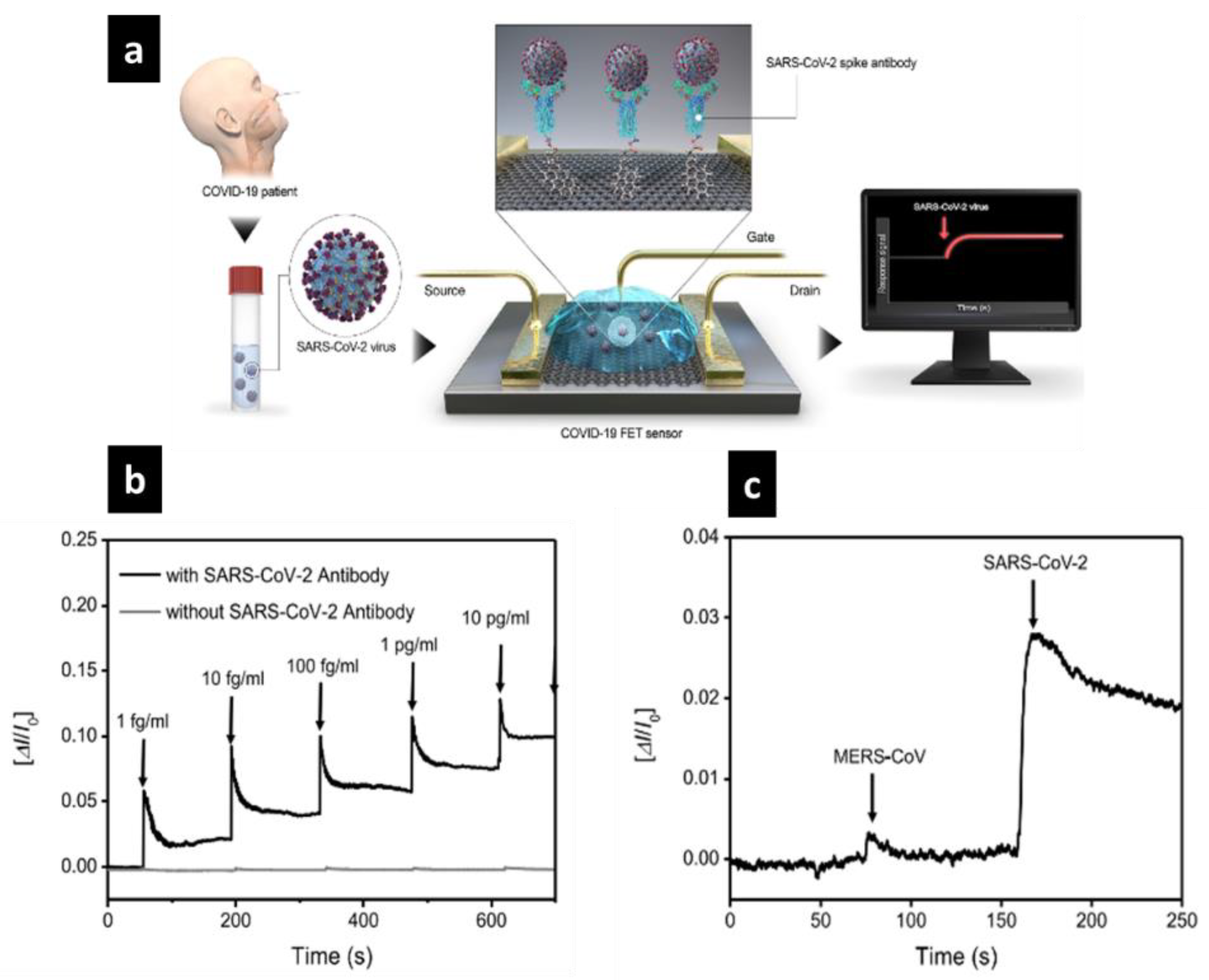
4.1.1. FET-Based Electrochemical Nanobiosensor
4.1.2. Cell-Based Electrochemical Nanobiosensor
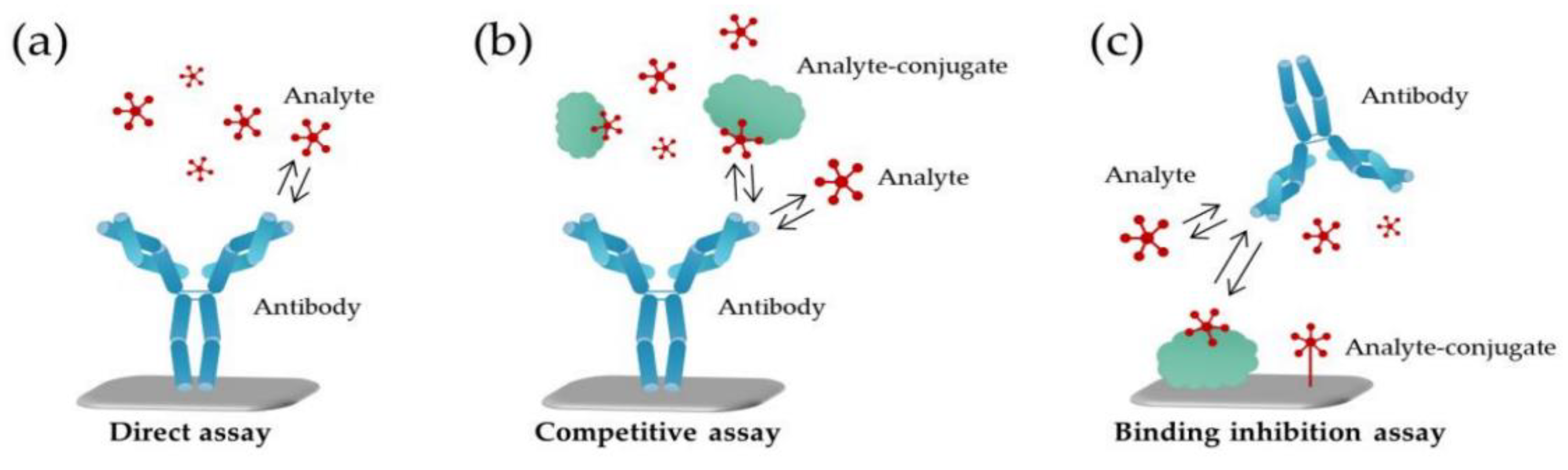
4.2. Optical Nanobiosensors
4.2.1. Magneto-Optical Nanobiosensors
4.2.2. Recently Developed COVID-19 Optical Nanobiosensors
4.3. Piezoelectric Nanobiosensors
5. Challenges and Opportunities for COVID-19 Causative Virus Nanosensors
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
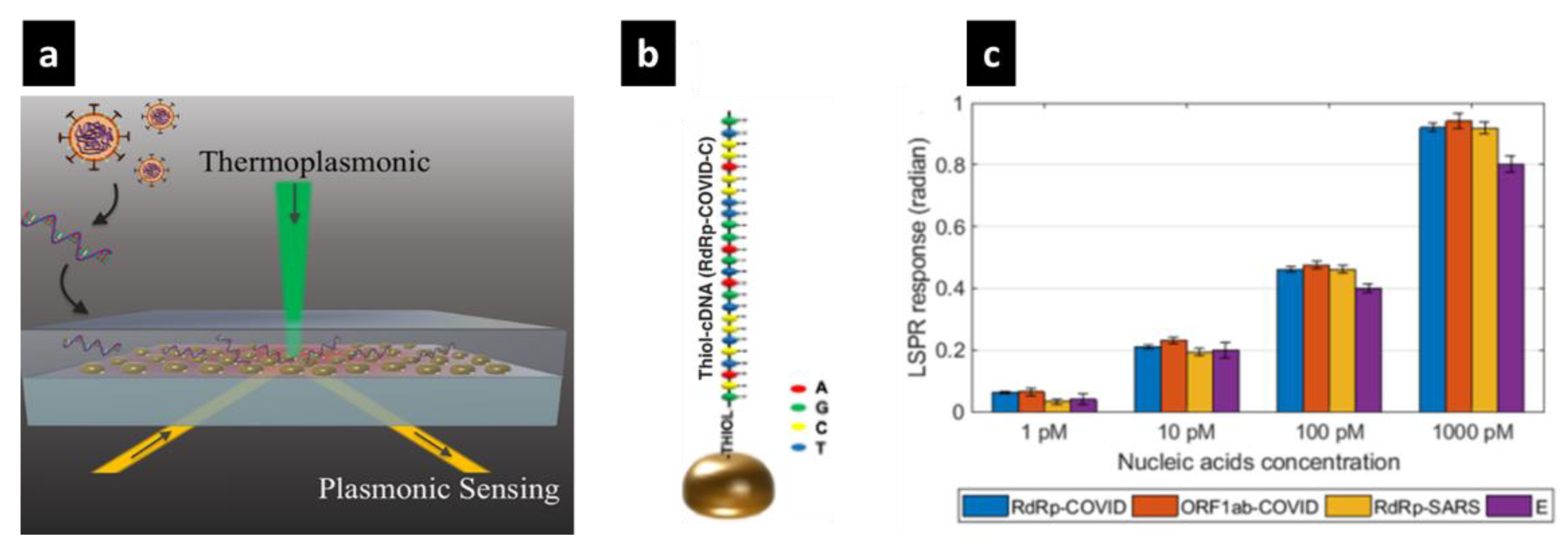
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhalla, N.; Pan, Y.; Yang, Z.; Payam, A.F. Opportunities and challenges for biosensors and nanoscale analytical tools for pandemics: Covid-19. ACS Nano 2020, 14, 7783–7807. [Google Scholar] [CrossRef] [PubMed]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef] [PubMed]
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121. [Google Scholar] [CrossRef]
- Palomar, Q.; Xu, X.; Gondran, C.; Holzinger, M.; Cosnier, S.; Zhang, Z. Voltammetric sensing of recombinant viral dengue virus 2 ns1 based on au nanoparticle–decorated multiwalled carbon nanotube composites. Microchim. Acta 2020, 187, 1–10. [Google Scholar] [CrossRef]
- Szunerits, S.; Saada, T.N.; Meziane, D.; Boukherroub, R. Magneto-Optical Nanostructures for Viral Sensing. Nanomaterials 2020, 10, 1271. [Google Scholar] [CrossRef]
- Asif, M.; Ajmal, M.; Ashraf, G.; Muhammad, N.; Aziz, A.; Iftikhar, T.; Wang, J.; Liu, H. The role of biosensors in coronavirus disease-2019 outbreak. Curr. Opin. Electrochem. 2020, 23, 174–184. [Google Scholar] [CrossRef]
- Rabi, F.A.; Al Zoubi, M.S.; Kasasbeh, G.A.; Salameh, D.M.; Al-Nasser, A.D. SARS-CoV-2 and Coronavirus Disease 2019: What We Know So Far. Pathogens 2020, 9, 231. [Google Scholar] [CrossRef]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ace2) in sars coronavirus–induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef]
- Jackson, J.K.; Weiss, M.A.; Schwarzenberg, A.B.; Nelson, R.M. Global Economic Effects of Covid-19. Available online: https://www.hsdl.org/?view&did=835306 (accessed on 1 September 2020).
- García-Magariño, I.; Varela-Aldas, J.; Palacios-Navarro, G.; Lloret, J. Fog computing for assisting and tracking elder patients with neurodegenerative diseases. Peer-To-Peer Netw. Appl. 2019, 12, 1225–1235. [Google Scholar] [CrossRef]
- Bilal, M.; Nazir, M.S.; Ahmed, I.; Iqbal, H. Coronaviruses and COVID-19–Complications and Lessons Learned for the Future. J. Pure Appl. Microbiol. 2020, 14, 725–731. [Google Scholar] [CrossRef]
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.-J.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (covid-19) outbreak–an update on the status. Military Med. Res. 2020, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, B.; Gao, F.; Fock, J.; Dufva, M.; Hansen, M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of sars-cov-2 rdrp coding sequence. Biosens. Bioelectron. 2020, 165, 112356. [Google Scholar] [CrossRef] [PubMed]
- Portela, F.; Oliveira, S.; Santos, M.; Machado, J.; Abelha, A. Ambient Intelligence for Health; A Real-Time Intelligent System for Tracking Patient Condition; Springer International Publishing: Cham, Switzerland, 2015; pp. 91–97. [Google Scholar]
- Mancini, F.; Barbanti, F.; Scaturro, M.; Errico, G.; Iacobino, A.; Bella, A.; Riccardo, F.; Marsili, G.; Stefanelli, P.; Pezzotti, P.; et al. Laboratory management for SARS-CoV-2 detection: A user-friendly combination of the heat treatment approach and rt-Real-time PCR testing. Emerg. Microbes Infect. 2020, 9, 1393–1396. [Google Scholar] [CrossRef]
- Chicaiza, F.A.; Lema-Cerda, L.; Marcelo Álvarez, V.; Andaluz, V.H.; Varela-Aldás, J.; Palacios-Navarro, G.; García-Magariño, I. Virtual Reality-Based Memory Assistant for the Elderly; Springer International Publishing: Cham, Switzerland, 2018; pp. 269–284. [Google Scholar]
- Vlădăreanu, L. Advanced intelligent control through versatile intelligent portable platforms. Sensors 2020, 20, 3644. [Google Scholar] [CrossRef]
- Ardeleanu, M.N.; Popescu, I.N.; Udroiu, I.N.; Diaconu, E.M.; Mihai, S.; Lungu, E.; Alhalaili, B.; Vidu, R. Novel PDMS-based Sensor System for MPWM Measurements of Picoliter Volumes in Microfluidic Devices. Sensors 2019, 19, 4886. [Google Scholar] [CrossRef] [Green Version]
- Cui, F.; Zhou, H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020, 165, 112349. [Google Scholar] [CrossRef]
- Kaya, S.I.; Karadurmus, L.; Ozcelikay, G.; Bakirhan, N.K.; Ozkan, S.A. Chapter 18—Electrochemical virus detections with nanobiosensors. In Nanosensors for Smart Cities; Han, B., Tomer, V.K., Nguyen, T.A., Farmani, A., Kumar Singh, P., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 303–326. [Google Scholar]
- Pandele, A.M.; Comanici, F.E.; Carp, C.A.; Miculescu, M.; Voicu, S.I.; Thakur, V.; Serban, B.C. Synthesis and characterization of cellulose acetate-hydroxyapatite micro and nano composites membranes for water purification and biomedical applications. Vacuum 2017, 146, 599–605. [Google Scholar] [CrossRef] [Green Version]
- Dziąbowska, K.; Czaczyk, E.; Nidzworski, D. Detection Methods of Human and Animal Influenza Virus—Current Trends. Biosensors 2018, 8, 94. [Google Scholar] [CrossRef] [Green Version]
- Koo, B.; Hong, K.H.; Jin, C.E.; Kim, J.Y.; Kim, S.-H.; Shin, Y. Arch-shaped multiple-target sensing for rapid diagnosis and identification of emerging infectious pathogens. Biosens. Bioelectron. 2018, 119, 79–85. [Google Scholar] [CrossRef]
- Kizek, R.; Krejcova, L.; Michalek, P.; Rodrigo, M.M.; Heger, Z.; Krizkova, S.; Vaculovicova, M.; Hynek, D.; Adam, V. Nanoscale virus biosensors: State of the art. Nanobiosens. Dis. Diagn. 2015, 4, 47–66. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, A.K.; Dev, A.; Karmakar, S. Nanosensors and nanobiosensors in food and agriculture. Environ. Chem. Lett. 2018, 16, 161–182. [Google Scholar] [CrossRef]
- Han, J.-H.; Lee, D.; Chew, C.H.C.; Kim, T.; Pak, J.J. A multi-virus detectable microfluidic electrochemical immunosensor for simultaneous detection of H1N1, H5N1, and H7N9 virus using ZnO nanorods for sensitivity enhancement. Sens. Actuators B Chem. 2016, 228, 36–42. [Google Scholar] [CrossRef]
- Han, K.N.; Li, C.A.; Bui, M.-P.N.; Pham, X.-H.; Kim, B.S.; Choa, Y.H.; Lee, E.K.; Seong, G.H. On-chip electrochemical detection of bio/chemical molecule by nanostructures fabricated in a microfluidic channel. Sens. Actuators B Chem. 2013, 177, 472–477. [Google Scholar] [CrossRef]
- Saylan, Y.; Denizli, A. Chapter 30—virus detection using nanosensors. In Nanosensors for Smart Cities; Han, B., Tomer, V.K., Nguyen, T.A., Farmani, A., Kumar Singh, P., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 501–511. [Google Scholar]
- Takemura, K.; Adegoke, O.; Takahashi, N.; Kato, T.; Li, T.-C.; Kitamoto, N.; Tanaka, T.; Suzuki, T.; Park, E.Y. Versatility of a localized surface plasmon resonance-based gold nanoparticle-alloyed quantum dot nanobiosensor for immunofluorescence detection of viruses. Biosens. Bioelectron. 2017, 89, 998–1005. [Google Scholar] [CrossRef] [PubMed]
- Palestino, G.; García-Silva, I.; González-Ortega, O.; Rosales-Mendoza, S. Can nanotechnology help in the fight against COVID-19? Expert Rev. Anti-Infect. Ther. 2020, 18, 849–864. [Google Scholar] [CrossRef]
- Talebian, S.; Wallace, G.G.; Schroeder, A.; Stellacci, F.; Conde, J. Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 2020, 15, 618–621. [Google Scholar] [CrossRef]
- Weiss, C.; Carriere, M.; Fusco, L.; Capua, I.; Regla-Nava, J.A.; Pasquali, M.; Scott, J.A.; Vitale, F.; Unal, M.A.; Mattevi, C.; et al. Toward Nanotechnology-Enabled Approaches against the COVID-19 Pandemic. ACS Nano 2020, 14, 6383–6406. [Google Scholar] [CrossRef]
- Rabiee, N.; Bagherzadeh, M.; Ghasemi, A.; Zare, H.; Ahmadi, S.; Fatahi, Y.; Dinarvand, R.; Rabiee, M.; Ramakrishna, S.; Shokouhimehr, M.; et al. Point-of-Use Rapid Detection of SARS-CoV-2: Nanotechnology-Enabled Solutions for the COVID-19 Pandemic. Int. J. Mol. Sci. 2020, 21, 5126. [Google Scholar] [CrossRef]
- Su, H.; Li, S.; Jin, Y.; Xian, Z.Y.; Yang, D.; Zhou, W.; Mangaran, F.; Leung, F.; Sithamparanathan, G.; Kerman, K. Nanomaterial-based biosensors for biological detections. Adv. Health Care Technol. 2017, 3, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Osman, B.; Uzun, L.; Beşirli, N.; Denizli, A. Microcontact imprinted surface plasmon resonance sensor for myoglobin detection. Mater. Sci. Eng. C 2013, 33, 3609–3614. [Google Scholar] [CrossRef] [PubMed]
- Bartold, K.; Pietrzyk-Le, A.; Golebiewska, K.; Lisowski, W.; Cauteruccio, S.; Licandro, E.; D’Souza, F.; Kutner, W. Oligonucleotide Determination via Peptide Nucleic Acid Macromolecular Imprinting in an Electropolymerized CG-Rich Artificial Oligomer Analogue. ACS Appl. Mater. Interfaces 2018, 10, 27562–27569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, S.; Hideshima, S.; Kuroiwa, S.; Nakanishi, T.; Osaka, T. Label-free detection of tumor markers using field effect transistor (FET)-based biosensors for lung cancer diagnosis. Sens. Actuators B Chem. 2015, 212, 329–334. [Google Scholar] [CrossRef]
- Erdem, Ö.; Saylan, Y.; Cihangir, N.; Denizli, A. Molecularly imprinted nanoparticles based plasmonic sensors for real-time Enterococcus faecalis detection. Biosens. Bioelectron. 2019, 126, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Anik, Ü.; Tepeli, Y.; Diouani, M.F. Fabrication of Electrochemical Model Influenza A Virus Biosensor Based on the Measurements of Neuroaminidase Enzyme Activity. Anal. Chem. 2016, 88, 6151–6153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.; Liu, W.; Zheng, M.; Wang, R. Label-Free and Ultrasensitive Electrochemical DNA Biosensor Based on Urchinlike Carbon Nanotube-Gold Nanoparticle Nanoclusters. Anal. Chem. 2020, 92, 4780–4787. [Google Scholar] [CrossRef]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar]
- Shandilya, R.; Bhargava, A.; Bunkar, N.; Tiwari, R.; Goryacheva, I.Y.; Mishra, P.K. Nanobiosensors: Point-of-care approaches for cancer diagnostics. Biosens. Bioelectron. 2019, 130, 147–165. [Google Scholar] [CrossRef]
- Pejcic, B.; De Marco, R.; Parkinson, G. The role of biosensors in the detection of emerging infectious diseases. Analyst 2006, 131, 1079–1090. [Google Scholar] [CrossRef]
- Ishikawa, F.N.; Chang, H.-K.; Curreli, M.; Liao, H.-I.; Olson, C.A.; Chen, P.-C.; Zhang, R.; Roberts, R.W.; Sun, R.; Cote, R.J.; et al. Label-Free, Electrical Detection of the SARS Virus N-Protein with Nanowire Biosensors Utilizing Antibody Mimics as Capture Probes. ACS Nano 2009, 3, 1219–1224. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Sun, Q.; He, J.; Xu, H.; Liu, C.; Zhao, C.; Xu, Y.; Wu, C.; Xiang, J.; Gu, D.; et al. Development of SPR biosensor for simultaneous detection of multiplex respiratory viruses. Bio-Med Mater. Eng. 2015, 26, S2207–S2216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Ai, S.; Chen, Q.; Yin, H.; Xu, J. Label-free electrochemical detection of Avian Influenza Virus genotype utilizing multi-walled carbon nanotubes–cobalt phthalocyanine–PAMAM nanocomposite modified glassy carbon electrode. Electrochem. Commun. 2009, 11, 1543–1546. [Google Scholar] [CrossRef]
- Xie, Z.; Huang, J.; Luo, S.; Xie, Z.; Xie, L.; Liu, J.; Pang, Y.; Deng, X.; Fan, Q. Ultrasensitive Electrochemical Immunoassay for Avian Influenza Subtype H5 Using Nanocomposite. PLoS ONE 2014, 9, e94685. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.H.; Shin, D.H.; Oh, J.; An, J.H.; Lee, J.S.; Jang, J. Multidimensional Conductive Nanofilm-Based Flexible Aptasensor for Ultrasensitive and Selective HBsAg Detection. ACS Appl. Mater. Interfaces 2018, 10, 28412–28419. [Google Scholar] [CrossRef] [PubMed]
- La Spada, L.; Vegni, L. Electromagnetic Nanoparticles for Sensing and Medical Diagnostic Applications. Materials 2018, 11, 603. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Jian, J.; Tu, T.; Yang, Z.; Ling, J.; Li, Y.; Wang, X.; Qiao, Y.; Tian, H.; Yang, Y.; et al. Wearable humidity sensor based on porous graphene network for respiration monitoring. Biosens. Bioelectron. 2018, 116, 123–129. [Google Scholar] [CrossRef]
- Adegoke, O.; Kato, T.; Park, E.Y. An ultrasensitive alloyed near-infrared quinternary quantum dot-molecular beacon nanodiagnostic bioprobe for influenza virus RNA. Biosens. Bioelectron. 2016, 80, 483–490. [Google Scholar] [CrossRef]
- Krejcova, L.; Nejdl, L.; Rodrigo, M.A.M.; Zurek, M.; Matousek, M.; Hynek, D.; Zitka, O.; Kopel, P.; Adam, V.; Kizek, R. 3D printed chip for electrochemical detection of influenza virus labeled with CdS quantum dots. Biosens. Bioelectron. 2014, 54, 421–427. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Ung, T.D.T.; Vu, T.H.; Tran, T.K.C.; Dong, V.Q.; Dinh, D.K.; Nguyen, Q.L. Fluorescence biosensor based on CdTe quantum dots for specific detection of H5N1 avian influenza virus. Adv. Nat. Sci. Nanosci. Nanotechnol. 2012, 3, 035014. [Google Scholar] [CrossRef]
- Shen, F.; Wang, J.; Xu, Z.; Wu, Y.; Chen, Q.; Li, X.; Jie, X.; Li, L.; Yao, M.; Guo, X.; et al. Rapid Flu Diagnosis Using Silicon Nanowire Sensor. Nano Lett. 2012, 12, 3722–3730. [Google Scholar] [CrossRef]
- Lin, C.-H.; Hung, C.-H.; Hsiao, C.-Y.; Lin, H.-C.; Ko, F.-H.; Yang, Y.-S. Poly-silicon nanowire field-effect transistor for ultrasensitive and label-free detection of pathogenic avian influenza DNA. Biosens. Bioelectron. 2009, 24, 3019–3024. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Karim, R.; Reda, Y.; Abdel-Fattah, A. Review—Nanostructured Materials-Based Nanosensors. J. Electrochem. Soc. 2020, 167, 03755. [Google Scholar] [CrossRef]
- Bezzon, V.D.N.; Montanheiro, T.L.D.A.; De Menezes, B.R.C.; Ribas, R.G.; Righetti, V.A.N.; Rodrigues, K.F.; Thim, G.P. Carbon Nanostructure-based Sensors: A Brief Review on Recent Advances. Adv. Mater. Sci. Eng. 2019, 2019, 4293073. [Google Scholar] [CrossRef] [Green Version]
- Alhalaili, B.; Dryden, D.M.; Vidu, R.; Ghandiparsi, S.; Cansizoglu, H.; Gao, Y.; Islam, M.S. High-aspect ratio micro- and nanostructures enabled by photo-electrochemical etching for sensing and energy harvesting applications. Appl. Nanosci. 2018, 8, 1171–1177. [Google Scholar] [CrossRef]
- Chen, Y.; Chan, K.-H.; Kang, Y.; Chen, H.; Luk, H.K.H.; Poon, R.W.S.; Chan, J.F.W.; Yuen, K.-Y.; Xia, N.; Lau, S.K.P.; et al. A sensitive and specific antigen detection assay for middle east respiratory syndrome coronavirus. Emerg. Microbes Infect. 2015, 4, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monajjemi, M.; Shahriari, S.; Mollaamin, F. Evaluation of Coronavirus Families & Covid-19 Proteins: Molecular Modeling Study. Biointerface Res. Appl. Chem. 2020, 10, 6039–6057. [Google Scholar]
- Layqah, L.A.; Eissa, S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta 2019, 186, 224. [Google Scholar] [CrossRef] [Green Version]
- Kakodkar, P.; Kaka, N.; Baig, M.N. A Comprehensive Literature Review on the Clinical Presentation, and Management of the Pandemic Coronavirus Disease 2019 (COVID-19). Cureus 2020, 12, e7560. [Google Scholar] [CrossRef] [Green Version]
- Junejo, Y.; Ozaslan, M.; Safdar, M.; Khailany, R.A.; Rehman, S.; Yousaf, W.; Khan, M.A. Novel SARS-CoV-2/COVID-19: Origin, pathogenesis, genes and genetic variations, immune responses and phylogenetic analysis. Gene Rep. 2020, 20, 100752. [Google Scholar] [CrossRef]
- Foster, P.L. Why Did the Flu Kill 80,000 Americans Last Year? Available online: https://theconversation.com/why-did-the-flu-kill-80-000-americans-last-year-105095 (accessed on 16 November 2020).
- Yusof, M.F.; Eltahir, Y.M.; Serhan, W.S.; Hashem, F.M.; Elsayed, E.A.; Marzoug, B.A.; Abdelazim, A.S.; Bensalah, O.K.A.; Al Muhairi, S.S. Prevalence of Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Abu Dhabi Emirate, United Arab Emirates. Virus Genes 2015, 50, 509–513. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Chen, L.; Tan, J.; Chen, J.; Du, L.; Sun, T.; Shen, J.; Chen, K.; Jiang, H.; Shen, X. Severe Acute Respiratory Syndrome Coronavirus 3C-like Proteinase N Terminus Is Indispensable for Proteolytic Activity but Not for Enzyme Dimerization. J. Biol. Chem. 2005, 280, 164–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besednova, N.N.; Zaporozhets, T.S.; Kuznetsova, T.A.; Makarenkova, I.D.; Fedyanina, L.N.; Kryzhanovsky, S.P.; Vishchuk, O.S.; Ermakova, S. Metabolites of Seaweeds as Potential Agents for the Prevention and Therapy of Influenza Infection. Mar. Drugs 2019, 17, 373. [Google Scholar] [CrossRef] [Green Version]
- Grifoni, A.; Sidney, J.; Zhang, Y.; Scheuermann, R.H.; Peters, B.; Sette, A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe 2020, 27, 671–680. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, J.; Bao, L.; Shi, Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020, 20, 398–400. [Google Scholar] [CrossRef]
- Acter, T.; Uddin, N.; Das, J.; Akhter, A.; Choudhury, T.R.; Kim, S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: A global health emergency. Sci. Total Environ. 2020, 730, 138996. [Google Scholar] [CrossRef]
- Bertram, S.; Heurich, A.; Lavender, H.; Gierer, S.; Danisch, S.; Perin, P.; Lucas, J.M.; Nelson, P.S.; Pöhlmann, S.; Soilleux, E.J. Influenza and SARS-Coronavirus Activating Proteases TMPRSS2 and HAT Are Expressed at Multiple Sites in Human Respiratory and Gastrointestinal Tracts. PLoS ONE 2012, 7, e35876. [Google Scholar] [CrossRef]
- Krejcova, L.; Hynek, D.; Michalek, P.; Milosavljevic, V.; Kopel, P.; Zitka, O.; Konecna, M.; Kynicky, J.; Adam, V.; Hubalek, J.; et al. Electrochemical Sensors and Biosensors for Influenza Detection–Literature Survey 2012–2013. Int. J. Electrochem. Sci. 2014, 9, 3440–3448. [Google Scholar]
- Zhang, H.; Miller, B.L. Immunosensor-based label-free and multiplex detection of influenza viruses: State of the art. Biosens. Bioelectron. 2019, 141, 111476. [Google Scholar] [CrossRef]
- Park, T.J.; Lee, S.J.; Kim, D.-K.; Heo, N.S.; Park, J.Y.; Lee, S.Y. Development of label-free optical diagnosis for sensitive detection of influenza virus with genetically engineered fusion protein. Talanta 2012, 89, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Wicklein, B.; Del Burgo, M.; Ángeles, M.; Yuste, M.; Carregal-Romero, E.; Llobera, A.; Darder, M.; Aranda, P.; Ortín, J.; Del Real, G.; et al. Biomimetic Architectures for the Impedimetric Discrimination of Influenza Virus Phenotypes. Adv. Funct. Mater. 2013, 23, 254–262. [Google Scholar] [CrossRef] [Green Version]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Singh, J.; Streithorst, J.; Granados, A.; Sotomayor-Gonzalez, A.; Zorn, K.; Gopez, A.; et al. Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-based DETECTR Lateral Flow Assay. medRxiv 2020. [Google Scholar] [CrossRef]
- Saylan, Y.; Yilmaz, F.; Özgür, E.; Derazshamshir, A.; Yavuz, H.; Denizli, A. Molecular Imprinting of Macromolecules for Sensor Applications. Sensors 2017, 17, 898. [Google Scholar] [CrossRef] [PubMed]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Saylan, Y.; Yılmaz, F.; Özgür, E.; Derazshamshir, A.; Bereli, N.; Yavuz, H.; Denizli, A. Surface plasmon resonance sensors for medical diagnosis. In Nanotechnology Characterization Tools for Biosensing and Medical Diagnosis; Kumar, C.S.S.R., Ed.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 425–458. [Google Scholar]
- Rim, T. Biosensors Based on Nanomaterials and Nanodevices [Book Review]. IEEE Nanotechnol. Mag. 2014, 8, 38. [Google Scholar] [CrossRef]
- Liang, K.-H.; Chang, T.-J.; Wang, M.-L.; Tsai, P.-H.; Lin, T.-H.; Wang, C.-T.; Yang, D.-M. Novel biosensor platforms for the detection of coronavirus infection and severe acute respiratory syndrome coronavirus 2. J. Chin. Med. Assoc. 2020, 83, 701–703. [Google Scholar]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.; Van Haperen, R.; Osterhaus, A.D.M.E.; Van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 1–6. [Google Scholar] [CrossRef]
- Siuzdak, K.; Niedziałkowski, P.; Sobaszek, M.; Łęga, T.; Sawczak, M.; Czaczyk, E.; Dziąbowska, K.; Ossowski, T.; Nidzworski, D.; Bogdanowicz, R. Biomolecular influenza virus detection based on the electrochemical impedance spectroscopy using the nanocrystalline boron-doped diamond electrodes with covalently bound antibodies. Sens. Actuators B Chem. 2019, 280, 263–271. [Google Scholar] [CrossRef]
- Miodek, A.; Sauriat-Dorizon, H.; Chevalier, C.; Delmas, B.; Vidic, J.; Korri-Youssoufi, H. Direct electrochemical detection of PB1-F2 protein of influenza A virus in infected cells. Biosens. Bioelectron. 2014, 59, 6–13. [Google Scholar] [CrossRef]
- Goode, J.A.; Rushworth, J.V.H.; Millner, P.A. Biosensor Regeneration: A Review of Common Techniques and Outcomes. Langmuir 2014, 31, 6267–6276. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Bhardwaj, A. Biosensor Technology for Pesticides—A review. Appl. Biochem. Biotechnol. 2015, 175, 3093–3119. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Cao, M.; Zhang, D.; Wang, Y.; Lu, Y.; Zhu, X.; Li, Y.; Xue, H.; Lin, Y.; Zhang, M.; Sun, Y.; et al. Clinical Features of Patients Infected with the 2019 Novel Coronavirus (COVID-19) in Shanghai, China. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Peltomaa, R.; Glahn-Martínez, B.; Benito-Peña, E.; Moreno-Bondi, M.C. Optical Biosensors for Label-Free Detection of Small Molecules. Sensors 2018, 18, 4126. [Google Scholar] [CrossRef] [Green Version]
- Borisov, S.M.; Wolfbeis, O.S. Optical biosensors. Chem. Rev. 2008, 108, 423–461. [Google Scholar] [CrossRef]
- Ribeiro, B.V.; Cordeiro, T.A.R.; e Freitas, G.R.O.; Ferreira, L.F.; Franco, D.L. Biosensors for the detection of respiratory viruses: A review. Talanta Open 2020, 2, 100007. [Google Scholar] [CrossRef]
- Sun, Z.; Ren, K.; Zhang, X.; Chen, J.; Jiang, Z.; Jiang, J.; Ji, F.; Ouyang, X.; Li, L. Mass spectrometry analysis of newly emerging coronavirus HCoV-19 spike S protein and human ACE2 reveals camouflaging glycans and unique post-translational modifications. Engineering 2020. [Google Scholar] [CrossRef]
- Roh, C.; Jo, S.K. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. Technol. Biotechnol. 2011, 86, 1475–1479. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-R.; Lee, G.-Y.; Chyi, J.-I.; Chang, C.-K.; Huang, C.-C.; Hsu, C.-P.; Huang, T.-H.; Ren, F.; Wang, Y.-L. Detection of Severe Acute Respiratory Syndrome (SARS) Coronavirus Nucleocapsid Protein Using AlGaN/GaN High Electron Mobility Transistors. ECS Trans. 2013, 50, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Kilianski, A.; Mielech, A.M.; Deng, X.; Baker, S.C. Assessing Activity and Inhibition of Middle East Respiratory Syndrome Coronavirus Papain-Like and 3C-Like Proteases Using Luciferase-Based Biosensors. J. Virol. 2013, 87, 11955–11962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teklemariam, A.D.; Samaddar, M.; Alharbi, M.G.; Al-Hindi, R.R.; Bhunia, A.K. Biosensor and molecular-based methods for the detection of human coronaviruses: A review. Mol. Cell. Probes 2020, 54, 101662. [Google Scholar] [CrossRef] [PubMed]
- Zuo, B.; Li, S.; Guo, Z.; Zhang, J.; Chen, C. Piezoelectric Immunosensor for SARS-Associated Coronavirus in Sputum. Anal. Chem. 2004, 76, 3536–3540. [Google Scholar] [CrossRef]
- Xie, B. Mini/micro thermal biosensors and other related devices for biochemical/clinical analysis and monitoring. TrAC Trends Anal. Chem. 2000, 19, 340–349. [Google Scholar] [CrossRef]
- Sinnarasa, I.; Thimont, Y.; Presmanes, L.; Barnabé, A.; Tailhades, P. Thermoelectric and Transport Properties of Delafossite CuCrO2:Mg Thin Films Prepared by RF Magnetron Sputtering. Nanomaterials 2017, 7, 157. [Google Scholar] [CrossRef] [Green Version]
- Llandro, J.; Palfreyman, J.J.; Ionescu, A.; Barnes, C.H.W. Magnetic biosensor technologies for medical applications: A review. Med. Biol. Eng. Comput. 2010, 48, 977–998. [Google Scholar] [CrossRef]
- Munawar, A.; Ong, Y.; Schirhagl, R.; Tahir, M.A.; Khan, W.S.; Bajwa, S.Z. Nanosensors for diagnosis with optical, electric and mechanical transducers. RSC Adv. 2019, 9, 6793–6803. [Google Scholar] [CrossRef] [Green Version]
- Ardakani, T.; Hosu, O.; Cristea, C.; Mazloum-Ardakani, M.; Marrazza, G. Latest Trends in Electrochemical Sensors for Neurotransmitters: A Review. Sensors 2019, 19, 2037. [Google Scholar] [CrossRef] [Green Version]
- Mubarok, A.Z.; Mani, V.; Huang, C.-H.; Chang, P.-C.; Huang, S.-T. Label-free electrochemical detection of neuraminidase activity: A facile whole blood diagnostic probe for infectious diseases. Sens. Actuators B Chem. 2017, 252, 641–648. [Google Scholar] [CrossRef]
- Hammond, J.L.; Formisano, N.; Estrela, P.; Carrara, S.; Tkac, J. Electrochemical biosensors and nanobiosensors. Essays Biochem. 2016, 60, 69–80. [Google Scholar] [PubMed] [Green Version]
- Ono, T.; Oe, T.; Kanai, Y.; Ikuta, T.; Ohno, Y.; Maehashi, K.; Inoue, K.; Watanabe, Y.; Nakakita, S.-I.; Suzuki, Y.; et al. Glycan-functionalized graphene-FETs toward selective detection of human-infectious avian influenza virus. Jpn. J. Appl. Phys. 2017, 56, 030302. [Google Scholar] [CrossRef]
- Chan, C.; Shi, J.; Fan, Y.; Yang, M. A microfluidic flow-through chip integrated with reduced graphene oxide transistor for influenza virus gene detection. Sens. Actuators B Chem. 2017, 251, 927–933. [Google Scholar] [CrossRef]
- Hassen, W.M.; Duplan, V.; Frost, E.; Dubowski, J.J. Quantitation of influenza A virus in the presence of extraneous protein using electrochemical impedance spectroscopy. Electrochim. Acta 2011, 56, 8325–8328. [Google Scholar] [CrossRef]
- Nidzworski, D.; Siuzdak, K.; Niedziałkowski, P.; Bogdanowicz, R.; Sobaszek, M.; Ryl, J.; Weiher, P.; Sawczak, M.; Wnuk, E.; Iii, W.A.G.; et al. A rapid-response ultrasensitive biosensor for influenza virus detection using antibody modified boron-doped diamond. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Hushegyi, A.; Pihíková, D.; Bertok, T.; Adam, V.; Kizek, R.; Tkac, J. Ultrasensitive detection of influenza viruses with a glycan-based impedimetric biosensor. Biosens. Bioelectron. 2016, 79, 644–649. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Li, Y.; Mao, X.; Huang, T.; Lu, H. Magnetic bio-nanobeads and nanoelectrode based impedance biosensor for detection of avian influenza virus. In Proceedings of the IEEE International Conference on Nano/Molecular Medicine and Engineering (NANOMED), Hong Kong, China, 5–9 December 2010; pp. 214–217. [Google Scholar]
- Lum, J.; Wang, R.; Lassiter, K.; Srinivasan, B.; Abi-Ghanem, D.; Berghman, L.; Hargis, B.; Tung, S.; Lu, H.; Li, Y. Rapid detection of avian influenza H5N1 virus using impedance measurement of immuno-reaction coupled with RBC amplification. Biosens. Bioelectron. 2012, 38, 67–73. [Google Scholar] [CrossRef]
- Fu, Y.; Callaway, Z.; Lum, J.; Wang, R.; Lin, J.; Li, Y. Exploiting Enzyme Catalysis in Ultra-Low Ion Strength Media for Impedance Biosensing of Avian Influenza Virus Using a Bare Interdigitated Electrode. Anal. Chem. 2014, 86, 1965–1971. [Google Scholar] [CrossRef]
- Singh, R.; Hong, S.; Jang, J. Label-free detection of influenza viruses using a reduced graphene oxide-based electrochemical immunosensor integrated with a microfluidic platform. Sci. Rep. 2017, 7, 42771. [Google Scholar] [CrossRef] [Green Version]
- Sayhi, M.; Ouerghi, O.; Belgacem, K.; Arbi, M.; Tepeli, Y.; Ghram, A.; Anik, Ü.; Österlund, L.; Laouini, D.; Diouani, M.F. Electrochemical detection of influenza virus h9n2 based on both immunomagnetic extraction and gold catalysis using an immobilization-free screen printed carbon microelectrode. Biosens. Bioelectron. 2018, 107, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, L.; Qiao, Z.; Lei, C.; Fu, Y.; Xie, Q.; Yao, S.; Li, Y.; Yingchun, F. Electrochemical Conversion of Fe3O4 Magnetic Nanoparticles to Electroactive Prussian Blue Analogues for Self-Sacrificial Label Biosensing of Avian Influenza Virus H5N1. Anal. Chem. 2017, 89, 12145–12151. [Google Scholar] [CrossRef] [PubMed]
- Veerapandian, M.; Hunter, R.; Neethirajan, S. Dual immunosensor based on methylene blue-electroadsorbed graphene oxide for rapid detection of the influenza A virus antigen. Talanta 2016, 155, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-H.; Zhuo, Y.; Yuan, R.; Chai, Y. A nanohybrid of platinum nanoparticles-porous ZnO–hemin with electrocatalytic activity to construct an amplified immunosensor for detection of influenza. Biosens. Bioelectron. 2016, 78, 321–327. [Google Scholar] [CrossRef]
- Lee, D.; Chander, Y.; Goyal, S.M.; Cui, T. Carbon nanotube electric immunoassay for the detection of swine influenza virus H1N1. Biosens. Bioelectron. 2011, 26, 3482–3487. [Google Scholar] [CrossRef]
- Anik, Ü.; Tepeli, Y.; Sayhi, M.; Nsiri, J.; Diouani, M.F. Towards the electrochemical diagnostic of influenza virus: Development of a graphene–Au hybrid nanocomposite modified influenza virus biosensor based on neuraminidase activity. Analyst 2018, 143, 150–156. [Google Scholar] [CrossRef]
- Bhardwaj, J.; Sharma, A.; Jang, J. Vertical flow-based paper immunosensor for rapid electrochemical and colorimetric detection of influenza virus using a different pore size sample pad. Biosens. Bioelectron. 2019, 126, 36–43. [Google Scholar] [CrossRef]
- Hai, W.; Goda, T.; Takeuchi, H.; Yamaoka, S.; Horiguchi, Y.; Matsumoto, A.; Miyahara, Y. Human influenza virus detection using sialyllactose-functionalized organic electrochemical transistors. Sens. Actuators B Chem. 2018, 260, 635–641. [Google Scholar] [CrossRef]
- Dong, S.; Zhao, R.; Zhu, J.; Lu, X.; Li, Y.; Qiu, S.; Jia, L.; Jiao, X.; Song, S.; Fan, C.; et al. Electrochemical DNA Biosensor Based on a Tetrahedral Nanostructure Probe for the Detection of Avian Influenza A (H7N9) Virus. ACS Appl. Mater. Interfaces 2015, 7, 8834–8842. [Google Scholar] [CrossRef]
- Ravina; Mohan, H.; Gill, P.S.; Kumar, A. Hemagglutinin gene based biosensor for early detection of swine flu (H1N1) infection in human. Int. J. Biol. Macromol. 2019, 130, 720–726. [Google Scholar] [CrossRef]
- Lee, T.; Park, S.Y.; Jang, H.; Kim, G.-H.; Lee, Y.; Park, C.; Mohammadniaei, M.; Lee, M.-H.; Min, J. Fabrication of electrochemical biosensor consisted of multi-functional DNA structure/porous au nanoparticle for avian influenza virus (H5N1) in chicken serum. Mater. Sci. Eng. C 2019, 99, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Diba, F.S.; Kim, S.; Lee, H.J. Amperometric bioaffinity sensing platform for avian influenza virus proteins with aptamer modified gold nanoparticles on carbon chips. Biosens. Bioelectron. 2015, 72, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Devarakonda, S.; Singh, R.; Bhardwaj, J.; Jang, J. Cost-Effective and Handmade Paper-Based Immunosensing Device for Electrochemical Detection of Influenza Virus. Sensors 2017, 17, 2597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarocka, U.; Sawicka, R.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Zagórski-Ostoja, W.; Sączyńska, V.; Porebska, A.J.; Dehaen, W.; Radecki, J.; et al. A biosensor based on electroactive dipyrromethene-Cu(II) layer deposited onto gold electrodes for the detection of antibodies against avian influenza virus type H5N1 in hen sera. Anal. Bioanal. Chem. 2015, 407, 7807–7814. [Google Scholar] [CrossRef] [PubMed]
- Mikuła, E.; Silva, C.E.; Kopera, E.; Zdanowski, K.; Radecki, J.; Radecka, H. Highly sensitive electrochemical biosensor based on redox—Active monolayer for detection of anti-hemagglutinin antibodies against swine-origin influenza virus H1N1 in sera of vaccinated mice. BMC Veter Res. 2018, 14, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malecka, K.; Świętoń, E.; Verwilst, P.; Stachyra, A.; Sirko, A.; Dehaen, W.; Radecki, J.; Radecka, H. Ultrasensitive electrochemical genosensor for direct detection of specific RNA sequences derived from avian influenza viruses present in biological samples. Acta Biochim. Pol. 2019, 66, 299–304. [Google Scholar] [CrossRef]
- Lee, H.E.; Kang, Y.; Choi, S. Electrochemical-DNA Biosensor Development Based on a Modified Carbon Electrode with Gold Nanoparticles for Influenza A (H1N1) Detection: Effect of Spacer. Int. J. Electrochem. Sci. 2014, 9, 6793–6808. [Google Scholar]
- Ishikawa, F.N.; Curreli, M.; Olson, C.A.; Liao, H.-I.; Sun, R.; Roberts, R.W.; Cote, R.J.; Thompson, M.E.; Zhou, C. Importance of Controlling Nanotube Density for Highly Sensitive and Reliable Biosensors Functional in Physiological Conditions. ACS Nano 2010, 4, 6914–6922. [Google Scholar] [CrossRef]
- Abad-Valle, P.; Fernández-Abedul, M.T.; Costa-García, A. Genosensor on gold films with enzymatic electrochemical detection of a SARS virus sequence. Biosens. Bioelectron. 2005, 20, 2251–2260. [Google Scholar] [CrossRef]
- De La Escosura-Muñiz, A.; González-Garcia, M.B.; Costa-García, A. DNA hybridization sensor based on aurothiomalate electroactive label on glassy carbon electrodes. Biosens. Bioelectron. 2007, 22, 1048–1054. [Google Scholar] [CrossRef]
- Martínez-Paredes, G.; González-García, M.B.; Costa-García, A. Genosensor for SARS Virus Detection Based on Gold Nanostructured Screen-Printed Carbon Electrodes. Electroanalysis 2009, 21, 379–385. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, F.; Xie, W.; Zhou, T.-C.; OuYang, J.; Jin, L.; Li, H.; Zhao, C.-Y.; Zhang, L.; Wei, J.; et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-COV-2 from the infected covid-19 patients using a smartphone. Sens. Actuators B Chem. 2021, 327, 128899. [Google Scholar] [CrossRef] [PubMed]
- Mahari, S.; Roberts, A.; Shahdeo, D.; Gandhi, S. eCovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 Antigen, a Spike Protein Domain 1 of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Laschi, S.; Mascini, M. Planar electrochemical sensors for biomedical applications. Med. Eng. Phys. 2006, 28, 934–943. [Google Scholar] [CrossRef]
- Bogomolova, A.; Komarova, E.; Reber, K.; Gerasimov, T.; Yavuz, O.; Bhatt, S.; Aldissi, M. Challenges of Electrochemical Impedance Spectroscopy in Protein Biosensing. Anal. Chem. 2009, 81, 3944–3949. [Google Scholar] [CrossRef] [PubMed]
- Janissen, R.; Sahoo, P.K.; Santos, C.A.; Da Silva, A.M.; Von Zuben, A.A.G.; Souto, D.E.P.; Costa, A.D.T.; Celedon, P.; Zanchin, N.I.T.; Almeida, D.B.; et al. InP Nanowire Biosensor with Tailored Biofunctionalization: Ultrasensitive and Highly Selective Disease Biomarker Detection. Nano Lett. 2017, 17, 5938–5949. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chen, X.; Wang, Q.; Xiao, M.; Zhong, D.; Sun, W.; Zhang, G.; Zhang, Z.-Y. Ultrasensitive Monolayer MoS2 Field-Effect Transistor Based DNA Sensors for Screening of Down Syndrome. Nano Lett. 2019, 19, 1437–1444. [Google Scholar] [CrossRef]
- Dai, Y.; Liu, C.-C. Recent Advances on Electrochemical Biosensing Strategies toward Universal Point-of-Care Systems. Angew. Chem. Int. Ed. 2019, 58, 12355–12368. [Google Scholar] [CrossRef]
- Chen, J.H.-K.; Yip, C.C.-Y.; Poon, R.W.-S.; Chan, K.-H.; Cheng, V.C.-C.; Hung, I.F.-N.; Chan, J.F.-W.; Yuen, K.-Y.; To, K.K.-W. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 1356–1359. [Google Scholar] [CrossRef]
- Choi, J.R. Development of Point-of-Care Biosensors for COVID-19. Front. Chem. 2020, 8, 517. [Google Scholar] [CrossRef]
- Cooper, D.R.; D’Anjou, B.; Ghattamaneni, N.; Harack, B.; Hilke, M.; Horth, A.; Majlis, N.; Massicotte, M.; Vandsburger, L.; Whiteway, E.; et al. Experimental Review of Graphene. ISRN Condens. Matter Phys. 2012, 2012, 1–56. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-H.; Park, S.-J.; Choi, J.-W. Electrical Property of Graphene and Its Application to Electrochemical Biosensing. Nanomaterials 2019, 9, 297. [Google Scholar] [CrossRef] [Green Version]
- Moschopoulou, G.; Kintzios, S. Application of “membrane-engineering” to bioelectric recognition cell sensors for the ultra-sensitive detection of superoxide radical: A novel biosensor principle. Anal. Chim. Acta 2006, 573, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Kokla, A.; Blouchos, P.; Livaniou, E.; Zikos, C.; Kakabakos, S.E.; Petrou, P.S.; Kintzios, S. Visualization of the membrane engineering concept: Evidence for the specific orientation of electroinserted antibodies and selective binding of target analytes. J. Mol. Recognit. 2013, 26, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Nabok, A.; Haron, S.; Ray, A. Planar silicon nitride waveguides for biosensing. IEE Proc. Nanobiotechnol. 2003, 150, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Quigley, G.R.; Harris, R.D.; Wilkinson, J.S. Sensitivity enhancement of integrated optical sensors by use of thin high-index films. Appl. Opt. 1999, 38, 6036–6039. [Google Scholar] [CrossRef] [PubMed]
- Luff, B.; Wilkinson, J.S.; Piehler, J.; Hollenbach, U.; Ingenhoff, J.; Fabricius, N. Integrated optical Mach-Zehnder biosensor. J. Light. Technol. 1998, 16, 583–592. [Google Scholar] [CrossRef]
- Malik, P.; Katyal, V.; Malik, V.; Asatkar, A.; Inwati, G.; Mukherjee, T.K. Nanobiosensors: Concepts and Variations. ISRN Nanomater. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- De Mol, N.J.; Fischer, M.J.E. Surface Plasmon Resonance: A General Introduction. In Methods in Molecular Biology: Surface Plasmon Resonance; De Mol, N.J., Fischer, M.J.E., Eds.; Humana Press: Totowa, NJ, USA, 2010; Volume 627, pp. 1–14. [Google Scholar]
- Daly, C.J.; McGrath, J.C. Fluorescent ligands, antibodies, and proteins for the study of receptors. Pharmacol. Ther. 2003, 100, 101–118. [Google Scholar] [CrossRef]
- Lee, B.; Roh, S.; Park, J. Current status of micro- and nano-structured optical fiber sensors. Opt. Fiber Technol. 2009, 15, 209–221. [Google Scholar] [CrossRef]
- Zhang, L.; Lou, J.; Tong, L. Micro/nanofiber optical sensors. Photon. Sens. 2010, 1, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Caygill, R.L.; Blair, G.E.; Millner, P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta 2010, 681, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Mutharasan, R. Review of biosensors for foodborne pathogens and toxins. Sens. Actuators B Chem. 2013, 183, 535–549. [Google Scholar] [CrossRef]
- Huang, J.C.; Chang, Y.-F.; Chen, K.-H.; Su, L.-C.; Lee, C.-W.; Chen, C.-C.; Chen, Y.-M.A.; Chou, C. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2009, 25, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Hayman, R. Fiber optic biosensors for bacterial detection. In Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Zourob, M., Elwary, S., Turner, A., Eds.; Springer International Publishing: Cham, Switzerland, 2008; pp. 125–137. [Google Scholar]
- Xu, S.; Ouyang, W.; Xie, P.; Lin, Y.; Qiu, B.; Lin, Z.; Chen, G.; Guo, L. Highly Uniform Gold Nanobipyramids for Ultrasensitive Colorimetric Detection of Influenza Virus. Anal. Chem. 2017, 89, 1617–1623. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Tsao, Y.-C.; Lee, F.-J.; Tsai, W.-H.; Wang, C.-H.; Chuang, T.-L.; Wu, M.-S.; Lin, C.-W. Optical fiber sensor based on surface plasmon resonance for rapid detection of avian influenza virus subtype H6: Initial studies. J. Virol. Methods 2016, 233, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.-T.; Bin Seo, H.; Kim, B.C.; Kim, S.K.; Song, C.-S.; Gu, M.B. Highly sensitive sandwich-type SPR based detection of whole H5Nx viruses using a pair of aptamers. Biosens. Bioelectron. 2016, 86, 293–300. [Google Scholar] [CrossRef]
- Wong, C.L.; Chua, M.; Mittman, H.; Choo, L.X.; Lim, H.Q.; Olivo, M. A Phase-Intensity Surface Plasmon Resonance Biosensor for Avian Influenza A (H5N1) Detection. Sensors 2017, 17, 2363. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.; Kim, G.H.; Kim, S.M.; Hong, K.; Kim, Y.; Park, C.; Sohn, H.; Min, J. Label-free localized surface plasmon resonance biosensor composed of multi-functional DNA 3 way junction on hollow Au spike-like nanoparticles (HAuSN) for avian influenza virus detection. Colloids Surfaces B Biointerfaces 2019, 182, 110341. [Google Scholar] [CrossRef]
- Chang, Y.-F.; Wang, W.-H.; Hong, Y.-W.; Yuan, R.-Y.; Chen, K.-H.; Huang, Y.-W.; Lu, P.-L.; Chen, Y.-H.; Chen, Y.-M.A.; Su, L.-C.; et al. Simple Strategy for Rapid and Sensitive Detection of Avian Influenza A H7N9 Virus Based on Intensity-Modulated SPR Biosensor and New Generated Antibody. Anal. Chem. 2018, 90, 1861–1869. [Google Scholar] [CrossRef]
- Zou, L.; Li, T.; Shen, R.; Ren, S.; Ling, L. A label-free light-up fluorescent sensing platform based upon hybridization chain reaction amplification and DNA triplex assembly. Talanta 2018, 189, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Rong, Z.; Wang, J.; Xiao, R.; Wang, S. A fluorescent aptasensor for H5N1 influenza virus detection based-on the core–shell nanoparticles metal-enhanced fluorescence (MEF). Biosens. Bioelectron. 2015, 66, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Guo, Y.; Zhao, M.; Sun, M.; Luo, F.; Guo, L.; Qiu, B.; Lin, Z.; Chen, G. Highly sensitive colorimetric immunosensor for influenza virus H5N1 based on enzyme-encapsulated liposome. Anal. Chim. Acta 2017, 963, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Luo, J.; Dong, W.; Wang, C.; Jin, W.; Xia, Y.; Wang, H.; Ding, H.; Jiang, L.; He, H. Development and evaluation of a polydiacetylene based biosensor for the detection of H5 influenza virus. J. Virol. Methods 2015, 219, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, L.; Wei, W.; Zhao, H.; Zhou, Z.; Zhang, Y.; Liu, S. Colorimetric detection of influenza A virus using antibody-functionalized gold nanoparticles. Analyst 2015, 140, 3989–3995. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Yang, S.C.; Choo, J. Early Diagnosis of Influenza Virus A Using Surface-enhanced Raman Scattering-based Lateral Flow Assay. Bull. Korean Chem. Soc. 2016, 37, 2019–2024. [Google Scholar] [CrossRef]
- Ye, W.W.; Tsang, M.-K.; Liu, X.; Yang, M.; Hao, J. Upconversion Luminescence Resonance Energy Transfer (LRET)-Based Biosensor for Rapid and Ultrasensitive Detection of Avian Influenza Virus H7 Subtype. Small 2014, 10, 2390–2397. [Google Scholar] [CrossRef]
- Park, T.J.; Hyun, M.S.; Lee, H.J.; Lee, S.Y.; Ko, S. A self-assembled fusion protein-based surface plasmon resonance biosensor for rapid diagnosis of severe acute respiratory syndrome. Talanta 2009, 79, 295–301. [Google Scholar] [CrossRef]
- Lee, S.J.; Park, J.P.; Park, T.J.; Lee, S.Y.; Lee, S.; Park, J.K. Selective Immobilization of Fusion Proteins on Poly(hydroxyalkanoate) Microbeads. Anal. Chem. 2005, 77, 5755–5759. [Google Scholar] [CrossRef]
- Waye, M.M.Y.; Law, P.; Wong, C.-H.; Au, T.; Chuck, C.-P.; Kong, S.-K.; Chan, P.; To, K.-F.; Lo, A.W.I.; Chan, J.Y.-W.; et al. The 3a Protein of SARS-coronavirus Induces Apoptosis in Vero E6 Cells. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Shanghai, China, 17–18 January 2006; pp. 7482–7485. [Google Scholar]
- Jang, K.J.; Lee, N.-R.; Yeo, W.S.; Jeong, Y.-J.; Kim, D.-E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008, 366, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Kim, Y.-K.; Kwon, H.-M.; Yeo, W.S.; Kim, D.-E.; Min, D.-H. A Graphene-Based Platform for the Assay of Duplex-DNA Unwinding by Helicase. Angew. Chem. Int. Ed. 2010, 49, 5703–5707. [Google Scholar] [CrossRef] [PubMed]
- Horejsh, D.; Martini, F.; Poccia, F.; Ippolito, G.; Di Caro, A.; Capobianchi, M.R. A molecular beacon, bead-based assay for the detection of nucleic acids by flow cytometry. Nucleic Acids Res. 2005, 33, e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, C.; Duan, J.-Z.; Wang, Z.-H.; Chen, Y.-Y.; Zhang, P.-H.; Zhan, L.; Yan, X.-Y.; Cao, W.-C.; Jin, G. Investigation of interaction between two neutralizing monoclonal antibodies and SARS virus using biosensor based on imaging ellipsometry. Biomed. Microdevices 2006, 8, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Park, T.J.; Lee, S.-K.; Yoo, S.M.; Yang, S.-M.; Lee, S.Y. Development of Reflective Biosensor Using Fabrication of Functionalized Photonic Nanocrystals. J. Nanosci. Nanotechnol. 2011, 11, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, M.; Hwang, J.; Kim, J.H.; Chung, D.-R.; Lee, K.-S.; Kang, M. Development of Label-Free Colorimetric Assay for MERS-CoV Using Gold Nanoparticles. ACS Sens. 2019, 4, 1306–1312. [Google Scholar] [CrossRef]
- Corman, V.M.; Müller, M.A.; Costabel, U.; Timm, J.; Binger, T.; Meyer, B.; Kreher, P.; Lattwein, E.; Eschbach-Bludau, M.; Nitsche, A.; et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Eurosurveillance 2012, 17, 20334. [Google Scholar] [CrossRef]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Multiplex Paper-Based Colorimetric DNA Sensor Using Pyrrolidinyl Peptide Nucleic Acid-Induced AgNPs Aggregation for Detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem. 2017, 89, 5428–5435. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Tian, D.; Liu, Y.; Lin, Z.; Lyon, C.J.; Lai, W.; Fusco, D.; Drouin, A.; Yin, X.; Hu, T.; et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020, 164, 112316. [Google Scholar] [CrossRef]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef]
- Kim, S.A.; Byun, K.M.; Kim, K.; Jang, S.M.; Ma, K.; Oh, Y.; Kim, D.; Kim, S.G.; Shuler, M.L.; Kim, S.J. Surface-enhanced localized surface plasmon resonance biosensing of avian influenza DNA hybridization using subwavelength metallic nanoarrays. Nanotechnology 2010, 21, 355503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, H.; Wang, R.; Hargis, B.M.; Lu, H.; Li, Y. A SPR Aptasensor for Detection of Avian Influenza Virus H5N1. Sensors 2012, 12, 12506–12518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saylan, Y.; Erdem, Ö.; Unal, S.; Denizli, A. An Alternative Medical Diagnosis Method: Biosensors for Virus Detection. Biosensors 2019, 9, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dafu, C.; Chen, X.; Wang, Y. Detection of SARS-CoV Antigen via SPR Analytical Systems with Reference. In Biosensors; Pier Andrea Serra, Ed.; InTechOpen: Rijeka, Croatia, 2010; pp. 169–178. [Google Scholar]
- Alimova, A.; Katz, A.; Podder, R.; Minko, G.; Wei, H.; Alfano, R.R.; Gottlieb, P. Virus Particles Monitored by Fluorescence Spectroscopy: A Potential Detection Assay for Macromolecular Assembly. Photochem. Photobiol. 2004, 80, 41–46. [Google Scholar] [CrossRef]
- Li, B.; Yu, Q.; Duan, Y. Fluorescent labels in biosensors for pathogen detection. Crit. Rev. Biotechnol. 2013, 35, 82–93. [Google Scholar] [CrossRef]
- Xi, Z.; Gong, Q.; Wang, C.; Zheng, B. Highly sensitive chemiluminescent aptasensor for detecting HBV infection based on rapid magnetic separation and double-functionalized gold nanoparticles. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Nie, R.; Huang, J.; Xu, X.; Yang, L. Immunoassays Using Optical-Fiber Sensor with All-Directional Chemiluminescent Collection. Anal. Chem. 2020, 92, 6257–6262. [Google Scholar] [CrossRef]
- Zeng, J.; Zhang, Y.; Zeng, T.; Aleisa, R.; Qiu, Z.; Chen, Y.; Huang, J.; Wang, D.; Yan, Z.; Yin, Y. Anisotropic plasmonic nanostructures for colorimetric sensing. Nano Today 2020, 32, 100855. [Google Scholar] [CrossRef]
- Rizal, C.; Pisana, S.; Hrvoic, I.; Fullerton, E. Microstructure and magneto-optical surface plasmon resonance of Co/Au multilayers. J. Phys. Commun. 2018, 2, 055010. [Google Scholar] [CrossRef]
- Rizal, C.; Pisana, S.; Hrvoic, I. Improved Magneto-Optic Surface Plasmon Resonance Biosensors. Photonics 2018, 5, 15. [Google Scholar] [CrossRef] [Green Version]
- Manera, M.G.; Colombelli, A.; Taurino, A.; Martin, A.G.; Rella, R. Magneto-Optical properties of noble-metal nanostructures: Functional nanomaterials for bio sensing. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brezoi, D.-V.; Ion, R.-M. Phase evolution induced by polypyrrole in iron oxide–polypyrrole nanocomposite. Sens. Actuators B Chem. 2005, 109, 171–175. [Google Scholar] [CrossRef]
- Armelles, G.; Cebollada, A.; García-Martín, A.; González, M.U. Magnetoplasmonics: Magnetoplasmonics: Combining Magnetic and Plasmonic Functionalities (Advanced Optical Materials 1/2013). Adv. Opt. Mater. 2013, 1, 2. [Google Scholar] [CrossRef]
- Brolo, A.G. Plasmonics for future biosensors. Nat. Photon. 2012, 6, 709–713. [Google Scholar] [CrossRef]
- Bonanni, V.; Bonetti, S.; Pakizeh, T.; Pirzadeh, Z.; Chen, J.; Nogués, J.; Vavassori, P.; Hillenbrand, R.; Åkerman, J.; Dmitriev, A. Designer Magnetoplasmonics with Nickel Nanoferromagnets. Nano Lett. 2011, 11, 5333–5338. [Google Scholar] [CrossRef]
- Rizal, C. Bio-Magnetoplasmonics, Emerging Biomedical Technologies and Beyond. J. Nanomed. Res. 2016, 3, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Takemura, K.; Park, E.Y. Plasmonic/magnetic graphene-based magnetofluoro-immunosensing platform for virus detection. Sens. Actuators B Chem. 2018, 276, 254–261. [Google Scholar] [CrossRef]
- Oh, S.; Kim, J.; Tran, V.; Lee, D.K.; Ahmed, S.R.; Hong, J.; Lee, J.; Park, E.Y.; Lee, J. Magnetic Nanozyme-Linked Immunosorbent Assay for Ultrasensitive Influenza A Virus Detection. ACS Appl. Mater. Interfaces 2018, 10, 12534–12543. [Google Scholar] [CrossRef]
- Trivedi, S.U.; Miao, C.; Sanchez, J.E.; Caidi, H.; Tamin, A.; Haynes, L.; Thornburg, N.J. Development and Evaluation of a Multiplexed Immunoassay for Simultaneous Detection of Serum IgG Antibodies to Six Human Coronaviruses. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Cui, H.; Song, W.; Ru, X.; Zhou, W.; Yu, X. A Simple Magnetic Nanoparticles-Based Viral RNA Extraction Method for Efficient Detection of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhanga, Q.; Liua, Y.; Niea, Y.; Ma, Q. Magnetic-plasmonic yolk-shell nanostructure-based plasmon-enhanced electrochemiluminescence sensor. Sens. Actuators B Chem. 2020, 319, 128245. [Google Scholar] [CrossRef]
- Li, M.-X.; Zhao, W.; Qian, G.-S.; Feng, Q.-M.; Xu, J.-J.; Chen, H.-Y. Distance mediated electrochemiluminescence enhancement of CdS thin films induced by the plasmon coupling of gold nanoparticle dimers. Chem. Commun. 2016, 52, 14230–14233. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.; Navale, G.R.; Dharne, M.S. Biosensors: Frontiers in rapid detection of COVID-19. 3 Biotech 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mauriz, E. Recent Progress in Plasmonic Biosensing Schemes for Virus Detection. Sensors 2020, 20, 4745. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Nie, Y.; Wang, M.; Zhang, Q.; Ma, Q. Distance-dependent plasmon-enhanced electrochemiluminescence biosensor based on MoS2 nanosheets. Biosens. Bioelectron. 2020, 148, 111823. [Google Scholar] [CrossRef]
- Bruinink, A.; Wang, J.; Wick, P. Effect of particle agglomeration in nanotoxicology. Arch. Toxicol. 2015, 89, 659–675. [Google Scholar] [CrossRef]
- Qiu, G.; Yue, Y.; Tang, J.; Zhao, Y.-B.; Wang, J. Total Bioaerosol Detection by a Succinimidyl-Ester-Functionalized Plasmonic Biosensor to Reveal Different Characteristics at Three Locations in Switzerland. Environ. Sci. Technol. 2020, 54, 1353–1362. [Google Scholar] [CrossRef]
- Deng, S.; Wang, P.; Yu, X. Phase-Sensitive Surface Plasmon Resonance Sensors: Recent Progress and Future Prospects. Sensors 2017, 17, 2819. [Google Scholar] [CrossRef] [Green Version]
- Dhama, K.; Khan, S.; Tiwari, R.; Sircar, S.; Bhat, S.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Coronavirus disease 2019–covid-19. Clin. Microbiol. Rev. 2020, 33, e00028-20. [Google Scholar] [CrossRef]
- Qiu, G.; Ng, S.-P.; Wu, C.-M.L. Bimetallic Au-Ag alloy nanoislands for highly sensitive localized surface plasmon resonance biosensing. Sens. Actuators B Chem. 2018, 265, 459–467. [Google Scholar] [CrossRef]
- Qiu, G.; Thakur, A.; Xu, C.; Ng, S.-P.; Lee, Y.; Wu, C.-M.L. Sensors/Biosensors: Detection of Glioma-Derived Exosomes with the Biotinylated Antibody-Functionalized Titanium Nitride Plasmonic Biosensor. Adv. Funct. Mater. 2019, 29, 1806761. [Google Scholar]
- Lee, J.-H.; Cheglakov, Z.; Yi, J.; Cronin, T.M.; Gibson, K.J.; Tian, B.; Weizmann, Y. Plasmonic Photothermal Gold Bipyramid Nanoreactors for Ultrafast Real-Time Bioassays. J. Am. Chem. Soc. 2017, 139, 8054–8057. [Google Scholar] [PubMed]
- Kim, M.; Lee, J.; Nam, J. Plasmonic Photothermal Nanoparticles for Biomedical Applications. Adv. Sci. 2019, 6, 1900471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Huang, P.; Nie, L.; Xing, R.; Liu, D.; Wang, Z.; Lin, J.; Chen, S.; Niu, G.; Lu, G.; et al. Single Continuous Wave Laser Induced Photodynamic/Plasmonic Photothermal Therapy Using Photosensitizer-Functionalized Gold Nanostars. Adv. Mater. 2013, 25, 3055–3061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Wang, L.; Callaway, Z.T.; Lu, H.; Huang, T.J.; Li, Y. A nanowell-based QCM aptasensor for rapid and sensitive detection of avian influenza virus. Sens. Actuators B Chem. 2017, 240, 934–940. [Google Scholar] [CrossRef] [Green Version]
- Erofeev, A.S.; Gorelkin, P.V.; Kolesov, D.V.; Kiselev, G.A.; Dubrovin, E.V.; Yaminsky, I.V. Label-free sensitive detection of influenza virus using PZT discs with a synthetic sialylglycopolymer receptor layer. R. Soc. Open Sci. 2019, 6, 190255. [Google Scholar]
- Wangchareansak, T.; Sangma, C.; Ngernmeesri, P.; Thitithanyanont, A.; Lieberzeit, P.A. Self-assembled glucosamine monolayers as biomimetic receptors for detecting WGA lectin and influenza virus with a quartz crystal microbalance. Anal. Bioanal. Chem. 2013, 405, 6471–6478. [Google Scholar] [CrossRef]
- Saylan, Y.; Akgönüllü, S.; Yavuz, H.; Unal, S.; Denizli, A. Molecularly Imprinted Polymer Based Sensors for Medical Applications. Sensors 2019, 19, 1279. [Google Scholar] [CrossRef] [Green Version]
- Prasad, S. Nanobiosensors: The future for diagnosis of disease? Nanobiosens. Disease Diagn. 2014, 3, 1–10. [Google Scholar]
- Yu, X.; Chen, F.; Wang, R.; Li, Y. Whole-bacterium SELEX of DNA aptamers for rapid detection of O157:H7 using a QCM sensor. J. Biotechnol. 2018, 266, 39–49. [Google Scholar] [CrossRef]
- Özgür, E.; Yılmaz, E.; Şener, G.; Uzun, L.; Say, R.; Denizli, A. A new molecular imprinting-based mass-sensitive sensor for real-time detection of 17β-estradiol from aqueous solution. Environ. Prog. Sustain. Energy 2012, 32, 1164–1169. [Google Scholar] [CrossRef]
- Wang, R.; Li, Y. Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens. Bioelectron. 2013, 42, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Diïltemiïz, S.E.; Ersöz, A.; Hür, D.; Keçili, R.; Say, R. 4-Aminophenyl boronic acid modified gold platforms for influenza diagnosis. Mater. Sci. Eng. C 2013, 33, 824–830. [Google Scholar]
- Hai, W.; Goda, T.; Takeuchi, H.; Yamaoka, S.; Horiguchi, Y.; Matsumoto, A.; Miyahara, Y. Specific Recognition of Human Influenza Virus with PEDOT Bearing Sialic Acid-Terminated Trisaccharides. ACS Appl. Mater. Interfaces 2017, 9, 14162–14170. [Google Scholar] [CrossRef] [PubMed]
- Owen, T.W.; Al-Kaysi, R.O.; Knöll, B.; Cheng, Q. Microgravimetric immunosensor for direct detection of aerosolized influenza A virus particles. Sens. Actuators B Chem. 2007, 126, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Tan, C.Y.; Tan, S.Y.; Wong, M.S.F.; Chen, Y.F.; Zhang, L.; Yao, K.; Gan, S.K.E.; Verma, C.; Tan, Y.-J. SAW sensor for Influenza A virus detection enabled with efficient surface functionalization. Sens. Actuators B Chem. 2015, 209, 78–84. [Google Scholar] [CrossRef]
- Yuan, Y.J.; Liang, T.; Han, K. Development of a Real-Time QCM Bond-Rupture System for POCT Applications. IEEE Sens. J. 2016, 16, 8731–8735. [Google Scholar] [CrossRef]
- Albano, D.; Shum, K.; Tanner, J.; Fung, Y. BS5.3—Piezoelectric quartz crystal aptamer biosensor for detection and quantification of SARS CoV helicase protein. In Proceedings of the 17th International Meeting on Chemical Sensors—IMCS 2018, Vienna, Austria, 15–19 July 2018; pp. 211–213. [Google Scholar]
- Pandey, L.M. Design of engineered surfaces for prospective detection of SARS-CoV-2 using quartz crystal microbalance-based techniques. Expert Rev. Proteom. 2020, 17, 425–432. [Google Scholar] [CrossRef]
- Hahm, J.-I. Biomedical Detection via Macro- and Nano-Sensors Fabricated with Metallic and Semiconducting Oxides. J. Biomed. Nanotechnol. 2013, 9, 1–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Disease | Flu (swine flu) | SARS 2002 | MERS 2012 | COVID 19 |
|---|---|---|---|---|
| Virus Name |  H1N1 Influenza A |  SARS–CoV-1 or SARS-CoV |  MERS-CoV |  SARS–CoV-2 or 2019-nCoV |
| Origin of Virus | Bird Influenza A | SARS-like BAT-CoV | SARS-like BAT-CoV | BaT-CoV RaTG13 or Pangolin-CoV |
| Intermediate host | Pig Influenza A | Civet Cat | Camel | Pangolin |
| Incubation period | 1–4 days | 2–7 days | 5 days | 2–14 days |
| Symptoms | Fever, cough, shortness of breath or difficulty breathing, mild respiratory tract infections, sometimes severe and acute diarrhea and acute vomiting | Fever, cough, shortness of breath or difficulty breathing, severe acute respiratory syndrome, 10 % mortality rate | Fever, cough, shortness of breath or difficulty breathing, severe acute respiratory syndrome, 37% mortality rate | Fever, cough, shortness of breath or difficulty breathing, severe acute respiratory syndrome; mortality rate; diarrhea, fatigue, vomiting, muscle or body aches, headache, loss of the sense of smell or taste. |
| Biomarkers | Normal Patient | Infected Patient | Severe Conditions | Ref. |
|---|---|---|---|---|
| Serum ferritin (ng/mL) | 15.0–150.0 | 800.4 (452.9–1451.6) | Inflammation | [90] |
| C-reactive protein (mg/mL) | 0.0–1.0 | 57.9 (20.9–103.2) | Viral infection | [90] |
| Interleukin-2R (U/mL) | 223.0–710.0 | 757.0 (528.5–1136.3) | High plasma concertation | [90,91] |
| Cytokines (IL-6) (pg/mL) | 0.0–7.0 | 7.9 | Syndrome related to “cytokine storm” | [92] |
| D-Dimer (µg/mL) | 0–0.243 | 0.5 | Abnormal blood coagulation distributed coagulopathy | [93] |
| Serum amyloid A (SAA) (mg/L) | 0–10 | 108.4 | Inflammation | [93] |
| Biological Samples | Nanomaterials | Detection Methods | Target | Limit of Detection (LOD) Linear Range (LR) | Ref. | |
|---|---|---|---|---|---|---|
| INFLUENZA VIRUSES | ||||||
| Biological substances such as DNA and proteins | Graphene (single-layer hexagonal carbon networks) | Field-Effect Transistor (FET) | Lectin | LOD: 130 pM LR: – | [110] | |
| Oligonucleotide sequences derived from an H5N1 avian influenza | rGO reduced graphene oxide | Field-Effect Transistor (FET) | Gene (H5N1) | LOD: 50 pM LR: – | [111] | |
| Saliva | Nanocrystalline boron-doped diamond | Electrochemical impedance (EIS) | Influenza virus M1 protein | LOD: 5 × 10−14 g/mL LR: – | [86] | |
| Egg sample | Graphene gold hybrid nanocomposite | Electrochemical impedance EIS) | Influenza A virus | LOD: 10–8 U/mL LR: 10−8–10−10 U/mL | [124] | |
| Fetal bovine serum, extraneous bovine serum albumin (BSA) | Gold electrode | Electrochemical impedance spectroscopy (EIS) | Human influenza virus type A (H3N2) | LOD: 8 ng/mL LR: | [112] | |
| Saliva buffer | Diamond biosensor (nano-scale boron-doped diamond surface sensor) | Electrochemical impedance spectroscopy (EIS) | M1 protein of influenza A virus | LOD: 1 fg/mL LR: 1–100 fg/mL | [113] | |
| Samples contained bovine serum albumin solution (BSA (0.5%) | Nanostructured hybrid bilayers on gold electrodes | Electrochemical impedance spectroscopy (EIS) | Human influenza A virus (H1N1) | LOD: 0.024 μg/mL LR: – | [78] | |
| Viral sample of inactivated, but intact influenza viruses H3N2 | Gold electrode | Electrochemical impedance spectroscopy (EIS) | Human influenza A virus (H3N2) | LOD: 1.3 × 104 viruses/mL LR: – | [114] | |
| Isolated AIV H5N1 sample incubated for 45 min at 37 °C | Magnetic iron oxide (Fe3O4) nanobeads | Electrochemical impedance spectroscopy (EIS) | Avian influenza virus (AIV) (H5N1) | LOD: 0.0128 HA unit/50 μL. LR: – | [116] | |
| Biological samples with avian influenza virus | Magnetic streptavidin-coated 30 nm nanobeads | Electrochemical impedance spectroscopy (EIS) | Avian influenza virus (AIV) (H5N1) | LOD: 103 EID 50/mL LR: – | [115] | |
| Inactivated avian influenza virus H5N1 sample | Concanavalin A-glucose oxidase-Au nanoparticles (ConA-GOx-AuNPs) | Electrochemical impedance spectroscopy (EIS) | Avian influenza virus (AIV) (H5N1) | LOD: 0.04 HAU/mL LR: – | [117] | |
| Commercial sample Spike saliva | Gold paper electrode | Electrochemical Impedance spectroscopy (EIS) | H1N1 antigen | LOD: 4.70 PFU/mL LR: – | [125] | |
| Influenza viral particles in infected swine nasal samples | Reduced graphene oxide nanosheets (RGO) | Chronoamperometry (CA) | Human influenza A virus (H1N1) | LOD: 0.5 PFU/mL LR: 1 to 104 PFU/mL | [118] | |
| Virus culture in embryonated chicken egg | Gold nanoparticles (AuNPs) | Chronoamperometry (CA) | Influenza virus (H9N2) | LOD: 16 HAU LR: – | [119] | |
| Virus samples in chicken embryo cultures | Conducting polymer of PEDOT-poly (3,4-ethylene-dioxythiophene) PSS film | Amperometry | Human influenza A virus (H1N1) | LOD: 0.025 HAU LR: – | [126] | |
| Commercial ELISA kits Probe sequence (avidin from egg whites) | ZnO-NRs Glass Wafer/PD MS | Amperometry | (H1N1), (H5N1), and (H7N9) influenza | LOD: 1.00 pg/mL LR: 1–10 ng/mL | [27] | |
| Throat swab samples | Gold electrode | Amperometry | Tetrahedral DNA probe of the H7N9/ssDNA of H7N9 | LOD: 0.750 pM LR: – | [127] | |
| Analyte samples collected from the throats of live animals, fecal content, and blood | Graphene oxide (GO) nanostructures Dual carbon SPE | Chronoamperometry and Differential pulse voltammetry (DPV) | HA proteins of Influenza virus (H5N1)/(H1N1) | LOD: 9.4 pM (Commercial H1N1) LOD: 8.3 pM (Commercial H5N1) LR: 25–100 pM | [121] | |
| Nasal swab and oropharyngeal samples | Gold screen printed electrode (SPE) | Cyclic voltammetry (CV) | ss-cDNA of the H1N1 | LOD: 0.004 ng in 6 µL | [128] | |
| Chicken serum | Gold electrode | Cyclic voltammetry (CV) | HA protein of H5N1 | LOD: 1.00 pM | [129] | |
| Negative chicken swab samples | Fe3O4 Magnetic Nanoparticles | Cyclic voltammetry (CV) | Avian influenza virus (AIV) (H5N1) | LOD: 0.367 HAU/mL LR: 0.0025 to 0.16 HAU | [120] | |
| Human blood, nasal swab, saliva, and urine | AP-Neu5Ac substrate | Cyclic Voltammetry (CV) Linear sweep Voltammetry (LSV) | Viral surface of NA-neuraminidase | LOD: 5.6 ng/mL LR: 0–900 nG/mL | [108] | |
| Cell culture and viral infection cells | Specific anti-PB1-F2 antibody on the surface of the Au micro-electrode modified with polypyrrole bearing ferrocene | Cyclic Voltammetry (CV) Differential pulse voltammetry (DPV) | Protein of influenza A virus (PB1-F2) | LOD: 0.42 nM LR: – | [87] | |
| Human serum | Pt NPs_porous ZnO spheres | Voltammetric (Cyclic Voltammetry) | DNA sequence of influenza virus | LOD: 0.76 pg/mL LR: 0.001–60 ng/mL | [122] | |
| Diluted human serum samples spiked | AuNPs | Differential pulse voltammetry (DPV) | H5N1 DNA aptamer/antiH5N1 | LOD: 100 fM LR: 100 fM–10 pM | [130] | |
| Saliva from a healthy person | Superhydrophobic paper/conductive carbon paste | Differential pulse voltammetry (DPV) | H1N1 antibody/H1N1 antigen | 113 PFU/mL | [131] | |
| Antibodies from Hen sera from individuals vaccinated and non-vaccinated | Gold electrode | Osteryoung square wave voltammetry (OSWV) | His6-H5 HA/antiH5N1 | LOD: 2.40 pg/mL LR: 4.0–100.0 pg/mL | [132] | |
| Diluted human and swine serum Vaccinated mice sera | Gold electrode | Osteryoung square wave voltammetry (OSWV). | His6-H1HA/anti-H1N1 | LOD: – LR: – | [133] | |
| Biological samples | Gold electrode | Osteryoung Square Wave Voltammetry (OSWV) | ssDNA of H5N1/ RNA of the H5N1 | LOD: 3.00 copies/µL | [134] | |
| The probe DNA (avidin-biotinylated probe DNA) | AuNPs | Voltammetric | Influenza virus type A (H1N1) | LOD: – LR: – | [135] | |
| Real patient samples | CdS quantum dots (QDs) | Voltammetric | Influenza virus | LOD: 0.06 mM LR: 0.06–0.5 mM | [53] | |
| Infected swine nasal samples | Single walled carbon nanotubes | Conductometry | Swine influenza virus (SIV) (H1N1) | LOD: 180 TCID50/mL LR: – | [123] | |
| Clinical exhaled breath condensate (EBC) samples | Silicon nanowire (SiNW) | Conductometry | Human influenza A viruses (H1N1) and (H3N2) | LOD: 2.9 viruses/µL LR: – | [55] | |
| Saliva from a healthy person | Superhydrophobic paper/conductive carbon paste | Differential pulse voltammetry (DPV) | H1N1 antibody/H1N1 antigen | 113 PFU/mL | [131] | |
| Antibodies from Hen sera from individuals vaccinated and non-vaccinated | Gold electrode | Osteryoung square wave voltammetry (OSWV) | His6-H5 HA/antiH5N1 | LOD: 2.40 pg/mL LR: 4.0–100.0 pg/mL | [132] | |
| CORONAVIRUSES | ||||||
| SARS-CoV | ||||||
| Streptavidin (S-Av) analyte | Single-walled carbon nanotubes (SWCNTs) | Field-Effect Transistor (FET) | nucleocapsid (N) protein of the SARS virus | LOD: physiological conditions | [136] | |
| Bovine serum albumin | In2O3 nanowire with an AMP (Fibronectin, Fn) | Field-Effect Transistor (FET) | SARS Virus N-Protein Nucleocapsid (N) | LOD: sub-nanomolar Concentration of N protein | [45] | |
| A 30-mer sequence of SARS Virus | 100 nm sputtered gold film | Cyclic voltammetry (CV) | SARS virus sequence | LOD: 7 × 10−6 M LR. 10−5 to 5 × 10−4 M | [137] | |
| Bovine serum albumin (BSA) and a rabbit immuno- Globulin G (RIgG) labeled with aurothiomalate | Au electrodeposition on glassy carbon electrodes (GCEs | Cyclic voltammetry (CV) | 30-mer sequence of the SARS virus | LOD: 15 fmol (30 μL) LR: – | [138] | |
| DNA sequence of SARS virus | Gold nanoparticles | Cyclic voltammetry (CV) Screen-printed carbon electrodes (SPCEs) | SARS virus sequence | LOD: 2.5 pmol/L LR: – | [139] | |
| Clinical specimens | Au@Fe3O4 nanocomposite Fe3O4 NPs (for premix A prep a-ration) and graphene oxide (GO) (for premix B preparation) | Differential pulse voltammetry (DPV) with a smartphone | RNA of SARS-CoV-2 | LOD: 200 copies/mL LR: – | [140] | |
| Protein sample of SARS-CoV | - | high electron mobility transistors (HEMTs | SARS-CoV nucleocapsid protein | LOD: 0.003 nM LR: – | [99] | |
| MERS-CoV or hCoV-EMC/2012 | ||||||
| Spiked nasal samples | AuNPs on carbon electrode | Square wave voltammetry (SWV) | Middle East respiratory syndrome coronavirus (MERS-CoV) and human coronavirus (hCoV) | LOD: 0.4 pg/mL LR: – | [62] | |
| 2019-nCoV or SARS-CoV-2 | ||||||
| Human Nasopharyngeal Swab Specimens, from COVID-19 patients Cultured virus | Graphene sheet | Field-Effect Transistor (FET) | SARS-CoV-2 spike protein | LOD: 1.6 × 101 pfu/mL in culture medium 2.42 × 102 copies/mL in clinical samples LR: – | [69] | |
| Green Monkey Kidney Cell Culture | Membrane-Engineered Vero Cells (Vero/Anti-S1) | Bioelectric Recognition Assay (BERA) | SARS-CoV-2 S1 Spike Protein Antigen | LOD: 1 fg/mL LR: – | [4] | |
| Spiked saliva samples | Fluorine doped tin oxide (FTO) electrode with gold nanoparticle (AuNPs) | Cyclic Voltammetry (CV), Differential Pulse Voltammetry (DPV) | nCovid-19 spike antigen (nCovid-19Ag) | LOD: 90 fM with eCovSens and 120 fM with potentiostat (spiked saliva samples LOD: 10 fM (in-house built device) of nCovid-19 Ag LR: 1 fM to 1 μM in standard buffer | [141] | |
| Biological Samples | Nanomaterials | Detection Methods | Target | Limit of Detection (LOD) Linear Range (LR) | Ref. |
|---|---|---|---|---|---|
| INFLUENZA VIRUSES | |||||
| H5N1 virus in biological samples | Gold nanoparticles (AuNPs) | Localized surface plasmon resonance (LSPR); Colorimetric | H5N1 virus | LOD: 0.086 mU/mL LR: 0.1–5 mU/mL | [165] |
| Viral strains, tracheal samples | Optical SPR fiber sensor | Surface plasmon resonance (SPR) | Avian Influenza virus | LOD: 5.14 × 105 EID50/0.1mL LR: – | [166] |
| H5N1–infected feces samples | Gold chip | Surface plasmon resonance (SPR) | H5N1 aptamer/H5N1 whole virus | LOD: 200 EID50/mL LR: – | [167] |
| Infected cells A549 type with wild type virus or with its PB1-F2 knock-out mutant | Immobilization of anti-PB1-F2 anti-body on the surface of Au micro-electrode modified with polypyrrole bearing ferrocene | Surface Plasmon Resonance (SPR) | PB1-F2 protein of influenza A virus | LOD: 0.42 nM LR: – | [87] |
| Biomolecular samples | Gold sensor | Surface plasmon resonance (SPR) | H5N1 antigen/H5N1 antibody ssDNA of the H1N1 | LOD: 193.3 ng/mL LR: – | [168] |
| Blood samples | Gold binding polypeptide (GBP)–fusion protein | Localized surface plasmon resonance/SPR imaging (LSPR/SPRi) | Influenza B virus | LOD: 1 pg/mL LR: – | [77] |
| Chicken serum | Au spike-like nanoparticle (hAuSN) immobilized on the indium-tin-oxide (ITO) substrate | Localized surface plasmon resonance (LSPR) | HA protein from H5N1 | LOD: 1.00 pM LR: – | [169] |
| Nasal mucosa from flulike syndrome patients | Gold chip | Intensity-modulated surface plasmon resonance (IM-SPR) | Attenuated reassorted H7N9 antigen | 402 copies/mL | [170] |
| Clinically isolated virus type H3N2 | Antibody-Gold nanoparticles | Fluorescence localized surface plasmon resonance (FL-LSPR) | H3N2 Virus | LOD: 10 PFU/mL LR: – | [30] |
| Human serum | DNA triplex with berberine | Fluorescence-fluorescein isothiocyanate assay (FL/FICT) | Gene of H7N9 virus DNA | LOD: 0.14 nM LR: – | [171] |
| Biological tissue | Quintenary alloyed CdZnSeTeS quantum dots | Near-infrared (NIR) Fluorescence | RNA sequence of influenza virus | LOD: 1 copy/mL LR: 0–14 copies/mL | [52] |
| Commercial H5N1–Human serum | Ag@SiO2 NPs | Fluorescence | H5N1 aptamer/Recombinant HA protein of H5N1 | LOD: 2.00–3.5 ng/mL LR: – | [172] |
| Human serum samples | Liposome-based sensor | Spectrophotometry | Influenza virus H5N1 based on enzyme encapsulated liposome | LOD: 0.04 ng/mL LR: 0.1–4.0 ng/mL | [173] |
| Tracheal swabs collected from wild birds | Polydiacetylene (PDA) vesicles | UV-VIS spectrometer | H5N1 antibody/HA of the H5N1 | LOD: 0.530 copies/µL LR: – | [174] |
| - | Gold nanoparticles (AuNPs) modified with monoclonal anti-hemagglutinin antibody (mAb). | Colorimetric immunosensor | Influenza A virus (IAV) | LOD: 7.8 hemagglutination units (HAU) LR: – | [175] |
| Viral culture | Gold nanoparticles (AuNPs) | Surface enhanced Raman scattering (SERS)-based lateral flow assay (LFA) | Viral particles (VP) | LOD: 1.9 × 104 PFU/mL LR: 0–1.0 × 106 PFU/mL | [176] |
| Viral nucleic acid | BaGdF 5: Yb/Er upconversion nanoparticles (UCNPs) to AuNPs | Luminescence Resonance Energy Transfer (LRET) | H7 hemagglutinin gene sequence | LOD: 7 pM LR: 10 pM to 10 nM | [177] |
| CORONAVIRUSES | |||||
| SARS-CoV | |||||
| Human serum Bovine serum albumin (BSA) | Gold nanoparticles | Localized surface plasmon coupled fluorescence (LSPCF) fiber-optic | SARS-CoV nucleocapsid protein (N protein) | LOD: 1 pg/mL LR: – | [163] |
| Rabbit anti-SCVme | Gold micropatterned chip | Surface plasmon resonance (SPR) | GBP-E-SCVme (SARS-CoV) fusion proteins/anti-SCVme | LOD: 0.200 µg/mL LR: – | [178] |
| Protein sample | – | Surface plasmon resonance (SPR) Fluorescence resonance energy transfer (FRET) | SARS-CoV genome sequence (full- length and N-terminal residues 1–7 deleted SARS 3Clpros) | LOD: – LR: – | [67] |
| Culture sample of SARS protein, enhanced GFP-green fluorescent protein and RFP-red fluorescent protein | poly(hydroxyalkanoate) (PHA) microbead | Fluorescence Flow cytometry | SARS-CoV envelope gene sequence | LOD: – LR: – | [179] |
| Vero E6 Cells | Green fluorescent protein (GFP) | Fluorescence | The 3a gene encodes a non-structural viral protein of SARS-Coronavirus | LOD. – LR: – | [180] |
| Protein sample | - | Fluorescence resonance energy transfer (FRET) | SARS coronavirus NTPase/Helicase | LOD: – LR: – | [181] |
| Upper-strand DNA and fluorescent-dye-conjugated bottom-strand DNA | Graphene oxide (GO) sheet | Fluorescence | SARS-CoV helicase | LOD: – LR: – | [182] |
| Lung samples cell (A549 human alveolar epithelial cells or inner medullary collecting duct (IMCD) mouse kidney epithelial cells taken after 6 days of infection with SARS-CoV) | - | Flow cytometry Affinity chromatography for purification of Spike-Fc protein) | SARS-CoV Spike Fc protein | LOD: – LR: – | [9] |
| Control samples Unlabeled nucleic acids in solution | - | Flow cytometry based on fluorescence | SARS-hCoV-M SARS-hCoV-N parainfluenza virus type 3(PIV-3), respiratory syncytial virus (RSV) | LOD: 26 fmol at an mean fluorescence intensity (MFI) of 5.7 (for SARS-N) LOD: 37 fmol (for SARS-M, hepatitis C virus , parainfluenza virus type 3, RSV) LR.26–56 fmol for SARS-M, HCV, PIV-3, RSV). | [183] |
| Serum samples (B cells of SARS convalescent patients; whole inactivated SARS-CoV virions) | Imaging ellipsometry (Real-time spectroscopic ellipsometry detect the protein layer pattern on the microarray surface. | B cells of SARS whole inactivated SARS-CoV virions | LOD: – LR: – | [184] | |
| Human serum from healthy donor Synthetic RNA aptamer | QDs-conjugated RNA aptamer On glass CHIP | optical QDs-based RNA aptamer chip | SARS-CoV N protein | LOD: concentrations as low as 0.1 pg/mL | [98] |
| genomic DNA | Functionalized Photonic Nanocrystals | Optical detection | SARS coronavirus antigenic surface protein | [185] | |
| Rabbit anti-SARS coronavirus surface antigen (Rabbit anti SCVme) | Gold micropatterned chip | Surface plasmon resonance (SPR) | SARS coronavirus surface antigen (SCVme) | LOD: 0.200 µg/mL LR: – | [178] |
| Serum samples (B cells of SARS convalescent patients; whole inactivated SARS-CoV virions) | Imaging ellipsometry (real-time spectroscopic ellipsometry detects the protein layer pattern on the microarray surface) | B cells of SARS whole inactivated SARS-CoV virions (scFv, b1 and h12 molecule) | LOD:2.2 µg/mL (b1) and 34 µg/mL (h12) LR: - | [184] | |
| Human serum from healthy donor Synthetic RNA aptamer | QDs-conjugated RNA aptamer On glass chip | Optical QDs-based RNA aptamer chip (Confocal laser scanning microscopy) | SARS-CoV N protein | LOD: concentrations as low as 0.1 pg ml−1 | [98] |
| MERS-CoV or hCoV-EMC/2012 | |||||
| Clinical sample | Gold nanoparticles (AuNPs) | Localized surface plasmon resonance (LSPR); Colorimetric assay | MERS-CoV DNA samples | LOD: 1 pmol/µL LR: – | [186] |
| Convalescent patient serum clinical isolate hCoV-EMC/2012 from green monkey kidney (Vero B4) cells | - | Immunofluorescence microscopy | hCoV-EMC/2012 (MERS-CoV) | LOD: 4.1 RNA copies/reaction | [187] |
| Synthetic DNA oligonucleotides samples | Silver nanoparticles (AgNPs) | Colorimetric assay | MERS-CoV DNA | LOD: 1.53 nM | [188] |
| 2019-nCoV or SARS-CoV-2 | |||||
| Respiratory secretions upper respiratory tract (URT) specimen | Gold nanoislands functionalized (AuNIs) with complementary DNA receptors | Plasmonic photo-thermal (PPT) and localized surface plasmon resonance (LSPR) | SARS-CoV-2 Nucleic acid | LOD: 0.22 pM LR: – | [1] |
| Clinical samples | - | Fluorescent detection | SARS-CoV-2 RNA | LOD: 2 copies per sample | [189] |
| Isolated RNA samples | Gold nanoparticles | Colorimetric assay | RNA sequence of SARS-CoV-2 | LOD: 0.18 ng/µL of RNA Dynamic range: 0.2–3 ng/µL. | [190] |
| Blood samples collected from 397 PCR confirmed COVID-19 patients and 128 negative patients | gold nanoparticle (AuNP) colloids | colorimetric assay | SARS-CoV-2 IgG-IgM combined antibody | LOD: – LR: – | [191] |
| Biological Samples | Nano-/Micro Materials | Detection Methods | Target | Limit of Detection (LOD) Linear Range (LR) | Ref. |
|---|---|---|---|---|---|
| INFLUENZA VIRUSES | |||||
| Virus samples in aqueous buffer and human serum | Ag@SiO2 nanoparticles | Metal enhanced fluorescence (MEF) | Influenza H5N1 | LOD: 3.5 ng/mL LR: 2–200 ng/mL | [172] |
| Clinical virus in complex biological samples | Au/Fe3O4 decorated graphene | Fluorescence | Influenza H1N1 | LOD: 7.27 fg/mL LR: 10–104 fg/mL | [209] |
| Complex biological samples | Au/iron oxides (Au/IONPs)-decorated graphene | Magnetofluoro immunoassay (Plasmonic-magnetic graphene platform for virus detection) | Influenza H1N1 In serum | LOD: 6.07 pg/mL LR: – | [209] |
| Clinically isolated human serum samples | Silica-shelled magnetic nanobeads (MagNBs) and gold nanoparticles | Magnetic nano(e)zyme-linked immunosorbent assay (MagLISA) | Influenza virus A | LOD: 5 × 10−12 g/mL (by human eyes) LOD: 44.2 × 10−15 g/mL (by a microplate reader) LR: 5 × 10−15–5 × 10−6 g/mL | [210] |
| CORONAVIRUSES | |||||
| SARS/MERS-CoV | |||||
| Paired human sera and control serum samples for each hCoV | Multiplexed magnetic microsphere | MMIA- multiplexed magnetic microsphere immunoassay Fluorescence | SARS-CoV and MERS-CoV Immunoglobulin G antibodies specific for recombinant nucleocapsid proteins (from SARS-CoV, and MERS-CoV, hCoVs, 229E, NL63, OC43, HKU1 | LOD: – LR: – | [211] |
| 2019-nCoV or SARS-CoV-2 | |||||
| SARS-CoV-2 pseudovirus in 200 μL serum samples | Poly (amino ester) with carboxyl groups (PC)-coated magnetic nanoparticle (pcMNPs) | Fluorescence and convectional RT-PCR protocol | Viral RNA extraction of SARS-CoV-2 | LOD: 10 copies of pseudovirus | [212] |
| Fetal bovine serum (FBS) | Magnetic nanoparticle (MNP) | Optomagnetic sensing | SARS-CoV-2 RdRp coding sequences | LOD: 0.4 fM dynamic Detection range: 3 orders of magnitude and a total assay time of ca. 100 min | [14] |
| Biological Samples | Nanomaterials | Detection Methods | Target | Limit of Detection (LOD) Linear Range (LR) | Ref. |
|---|---|---|---|---|---|
| INFLUENZA VIRUSES | |||||
| - | Gold film | Quartz crystal microbalance (QCM) | Hemagglutinin (HA) protein of influenza A virus | LOD: 4.7 × 10−2 µM, (0.26 µg/mL) LR: – | [235] |
| Sample of human influenza A virus (H1N1) incubated in a chicken egg culture | Poly(EDOT-co- EDOTOA) Films | Quartz crystal microbalance (QCM) | Human influenza A virus H1N1 | LOD: 0.012 HAU LR: – | [236] |
| Influenza A virus (VR-544, H3N2) samples | QCM electrodes | Quartz crystal microbalance (QCM) | Influenza A virions, influenza H3N2 polyclonal IgG | LOD: 4 virus particles/mL | [237] |
| Commercial H5N3 | Lead zirconate titanate (PZT) piezoelectric disc | Piezoelectric–SPM | H5N3 surface glycoprotein | 105 vp/mL (100 µm thick) | [228] |
| Biological sample | – | Surface acoustic wave (SAW) | HA proteins of Influenza A virus sub type H1N1 | LOD: 1 ng /mL LR: – | [238] |
| commercial H5N3 | Lead zirconate titanate (PZT) piezoelectric disc | Piezoelectric—SPM | 3′SLPAA polymer/H5N3 surface glycoprotein | 105 vp/mL (100 µm thick) | [228] |
| CORONAVIRUSES | |||||
| SARS-CoV | |||||
| Sputum | PZ crystal surface | Immunoassay | SARS-CoV | LOD: 0.6 µg/mL LR: – | [102] |
| Biological sample | Piezoelectric immunosensor | Quartz crystal microbalance (QCM) | SARS-CoV spike protein S1 | – | [239] |
| High protein sera sample | Aptamer coated paramagnetic nanoparticles | Piezoelectric | SARS-CoV helicase protein | LOD: 3.5 ng/mL | [240] |
| 2019-nCoV or SARS-CoV-2 | |||||
| Oral swab samples | Nanoparticles | Quartz crystal microbalance (QCM) | SARS-CoV-2 spike protein | – | [241] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhalaili, B.; Popescu, I.N.; Kamoun, O.; Alzubi, F.; Alawadhia, S.; Vidu, R. Nanobiosensors for the Detection of Novel Coronavirus 2019-nCoV and Other Pandemic/Epidemic Respiratory Viruses: A Review. Sensors 2020, 20, 6591. https://doi.org/10.3390/s20226591
Alhalaili B, Popescu IN, Kamoun O, Alzubi F, Alawadhia S, Vidu R. Nanobiosensors for the Detection of Novel Coronavirus 2019-nCoV and Other Pandemic/Epidemic Respiratory Viruses: A Review. Sensors. 2020; 20(22):6591. https://doi.org/10.3390/s20226591
Chicago/Turabian StyleAlhalaili, Badriyah, Ileana Nicoleta Popescu, Olfa Kamoun, Feras Alzubi, Sami Alawadhia, and Ruxandra Vidu. 2020. "Nanobiosensors for the Detection of Novel Coronavirus 2019-nCoV and Other Pandemic/Epidemic Respiratory Viruses: A Review" Sensors 20, no. 22: 6591. https://doi.org/10.3390/s20226591
APA StyleAlhalaili, B., Popescu, I. N., Kamoun, O., Alzubi, F., Alawadhia, S., & Vidu, R. (2020). Nanobiosensors for the Detection of Novel Coronavirus 2019-nCoV and Other Pandemic/Epidemic Respiratory Viruses: A Review. Sensors, 20(22), 6591. https://doi.org/10.3390/s20226591





