Characterization of Antibiotic Resistance in Select Tertiary Hospitals in Uganda: An Evaluation of 2020 to 2023 Routine Surveillance Data
Abstract
:1. Background
2. Methodology
2.1. Sample Size
2.2. Statistical Analysis
2.3. Ethical Statement
3. Results
3.1. Susceptibility of Isolates in General and Specialized Units
3.2. Antibiotic Susceptibility Variation by Hospital Units
3.3. Antibiotic Susceptibility Variation by Age Group
3.4. Antibiotic Susceptibility Variation by Sex
3.5. Antibiotic Susceptibility Variation by Sample Type
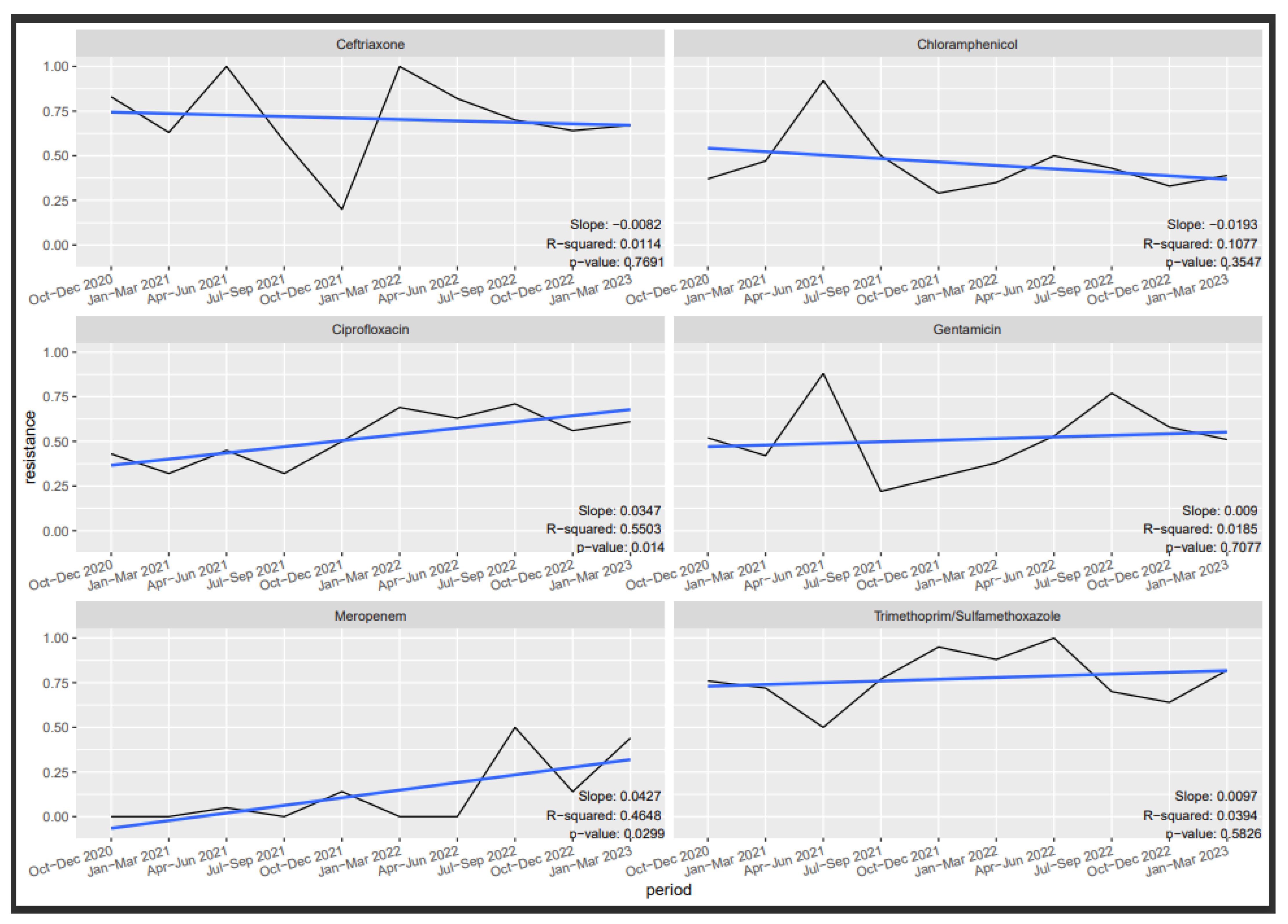
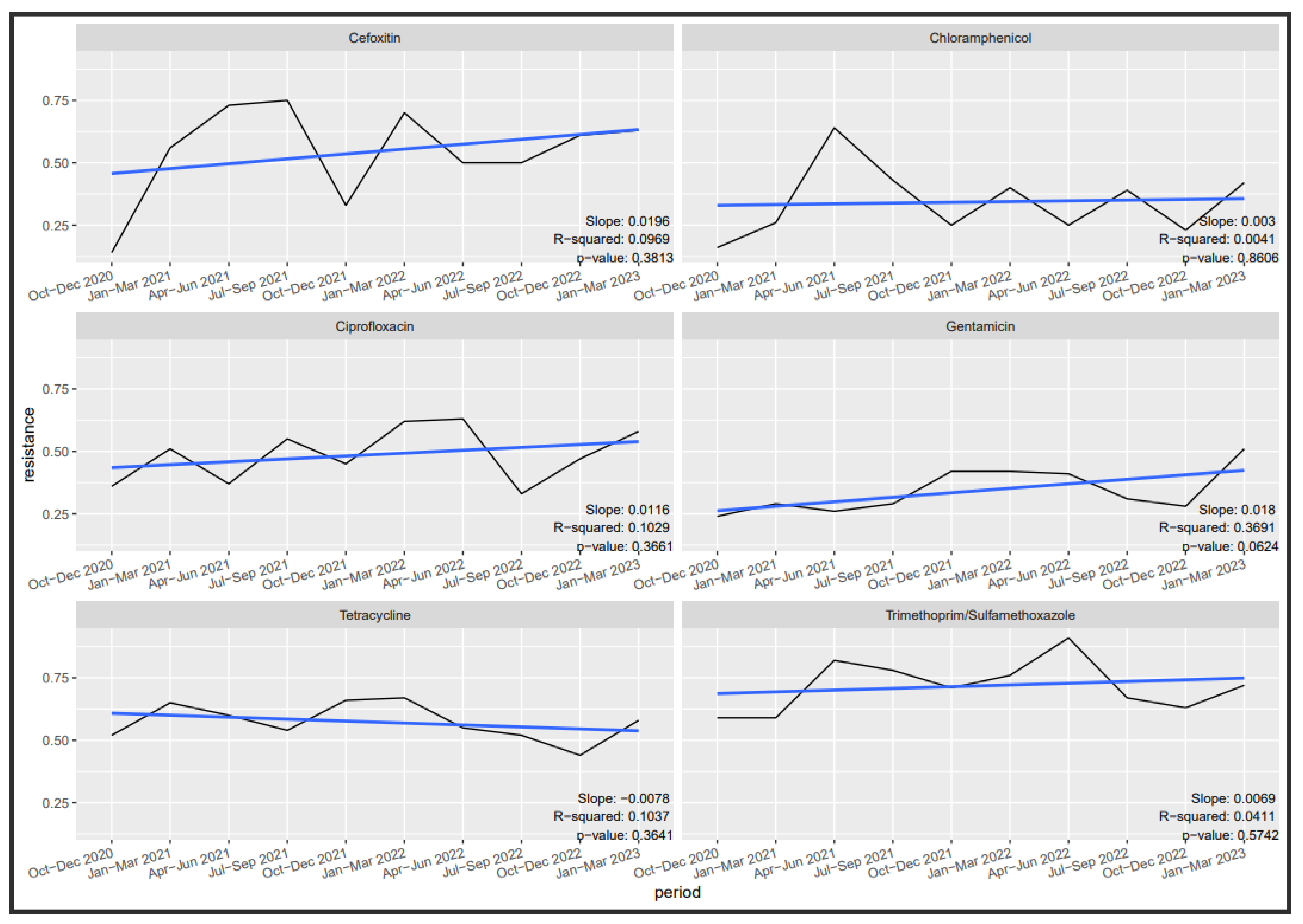
3.6. Antibiotic Susceptibility Trends
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial Resistance |
| AWaRe | Access Watch Reserve |
| AST | Antimicrobial Susceptibility Testing |
| GLASS | Global Antimicrobial Resistance and Use Surveillance System |
| CRE | Carbapenem Resistant Enterobacterales |
| ESBL | Extended Spectrum Beta Lactamases |
| MDR | Multi-Drug Resistance |
| MRSA | Methicillin Resistant Staphylococcus Aureus |
| NAP | National Action Plan |
| SLMTA | Strengthening Laboratory Management Towards |
| Accreditation | |
| SLIPTA | Laboratory Quality Improvement Process Towards |
| VRE | Vancomycin Resistant Enterococcus |
References
- Jim, O. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. 2014. Available online: https://wellcomecollection.org/works/rdpck35v (accessed on 4 March 2024).
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Elton, L.; Thomason, M.J.; Tembo, J.; Velavan, T.P.; Pallerla, S.R.; Arruda, L.B.; Vairo, F.; Montaldo, C.; Ntoumi, F.; Hamid, M.M.A.; et al. Antimicrobial resistance preparedness in sub-Saharan African countries. Antimicrob. Resist. Infect. Control 2020, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- Kateete, D.P.; Namazzi, S.; Okee, M.; Okeng, A.; Baluku, H.; Musisi, N.L.; A Katabazi, F.; Joloba, M.L.; Ssentongo, R.; Najjuka, F.C. High prevalence of methicillin resistant Staphylococcus aureus in the surgical units of Mulago hospital in Kampala, Uganda. BMC Res. Notes 2011, 4, 326. [Google Scholar] [CrossRef] [PubMed]
- Bebell, L.M.; Ngonzi, J.; Bazira, J.; Fajardo, Y.; Boatin, A.A.; Siedner, M.J.; Bassett, I.V.; Nyehangane, D.; Nanjebe, D.; Jacquemyn, Y.; et al. Antimicrobial-resistant infections among postpartum women at a Ugandan referral hospital. PLoS ONE 2017, 12, e0175456. [Google Scholar] [CrossRef] [PubMed]
- UNAS; CDDEP; GARP-Uganda; Mpairwe, Y.; Wamala, S. Antibiotic Resistance in Uganda: Situation Analysis and Recommendations; Uganda National Academy of Sciences; Center for Disease Dynamics, Economics & Policy: Kampala, Uganda, 2015; 107p. [Google Scholar]
- Nabadda, S.; Kakooza, F.; Kiggundu, R.; Walwema, R.; Bazira, J.; Mayito, J.; Mugerwa, I.; Sekamatte, M.; Kambugu, A.; Lamorde, M.; et al. Implementation of the World Health Organization Global Antimicrobial Resistance Surveillance System in Uganda, 2015–2020: Mixed-Methods Study Using National Surveillance Data. JMIR Public Health Surveill. 2021, 7, e29954. [Google Scholar] [CrossRef] [PubMed]
- Mugerwa, I.; Nabadda, S.N.; Midega, J.; Guma, C.; Kalyesubula, S.; Muwonge, A. Antimicrobial Resistance Situational Analysis 2019–2020: Design and Performance for Human Health Surveillance in Uganda. Trop. Med. Infect. Dis. 2021, 6, 178. [Google Scholar] [CrossRef]
- Mahon, C.R.; Lehman, D.C.; Manuselis, G. Textbook of Diagnostic Microbiology, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- CLSI. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data, 5th ed.; CLSI Guidelines M39; CLSI: Wayne, PA, USA, 2022. [Google Scholar]
- CLSI. Performance Standards Antimicrobial Susceptibility Testing, 33rd ed.; CLSI Supplement M100; CLSI: Wayne, PA, USA, 2023. [Google Scholar]
- CLSI. Performance Standards Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- WHONET ©. The Microbiology Laboratory Database Software. 1989–2023. Available online: https://whonet.org/ (accessed on 4 March 2024).
- Ampaire, L.; Muhindo, A.; Orikiriza, P.; Mwanga-Amumpaire, J.; Bebell, L.; Boum, Y. A review of antimicrobial resistance in East Africa. Afr. J. Lab. Med. 2016, 5, a432. [Google Scholar] [CrossRef]
- Seni, J.; Najjuka, C.F.; Kateete, D.P.; Makobore, P.; Joloba, M.L.; Kajumbula, H.; Kapesa, A.; Bwanga, F. Antimicrobial resistance in hospitalized surgical patients: A silently emerging public health concern in Uganda. BMC Res. Notes 2013, 6, 298. [Google Scholar] [CrossRef]
- Ongom, P.A.; Obuku, E.A.; Achan, B. Community-acquired soft-tissue pyogenic abscesses in Mulago hospital, Kampala: Bacteria isolated and antibiotic sensitivity. East Cent. Afr. J. Surg. 2012, 17, 28–37. [Google Scholar]
- Pius, T.; Irege, R.; Makeri, D.; Tamale, A. Methicillin-Resistant Staphylococcus aureus among Patients with Skin and Soft Tissue Infections: A Cross-Sectional Study at a Tertiary Hospital in Bushenyi, Western, Uganda. Open Access Libr. J. 2023, 10, e10186. [Google Scholar] [CrossRef]
- Wangai, F.K.; Masika, M.M.; Maritim, M.C.; Seaton, R.A. Methicillin-resistant Staphylococcus aureus (MRSA) in East Africa: Red alert or red herring? BMC Infect. Dis. 2019, 19, 596. [Google Scholar] [CrossRef]
- Kumburu, H.H.; Sonda, T.; Leekitcharoenphon, P.; van Zwetselaar, M.; Lukjancenko, O.; Alifrangis, M.; Lund, O.; Mmbaga, B.T.; Kibiki, G.; Aarestrup, F.M. Hospital Epidemiology of Methicillin-Resistant Staphylococcus aureus in a Tertiary Care Hospital in Moshi, Tanzania, as Determined by Whole Genome Sequencing. BioMed Res. Int. 2018, 2018, 2087693. [Google Scholar] [CrossRef]
- Georges, M.; Odoyo, E.; Matano, D.; Tiria, F.; Kyany’a, C.; Mbwika, D.; Mutai, W.C.; Musila, L. Determination of Enterococcus faecalis and Enterococcus faecium Antimicrobial Resistance and Virulence Factors and Their Association with Clinical and Demographic Factors in Kenya. J. Pathog. 2022, 2022, 3129439. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Publishes List of Bacteria for Which Now Antibiotics Are Urgently Needed. [Internet] 2022 Jan. Available online: https://www.ecdc.europa.eu/en/news-events/who-publishes-list-bacteria-which-new-antibiotics-are-urgently-needed#:~:text=WHO%20publishes%20list%20of%20bacteria%20for%20which%20new%20antibiotics%20are%20urgently%20needed,-News&text=%E2%80%8BThe%20World%20Health%20Organization,effective%20antibiotics%20are%20urgently%20needed (accessed on 4 March 2024).
- President, J.C.; De Waele, J. Intensive Care Units as Epicentres for Antimicrobial Resistance Development. AMR Control 2017. Available online: http://resistancecontrol.info/2017/intensive-care-units-as-epicentres-for-antimicrobial-resistance-development/ (accessed on 4 March 2024).
- Yildirim, S.; Nursal, T.Z.; Tarim, A.; Torer, N.; Noyan, T.; Demiroglu, Y.Z.; Moray, G.; Haberal, M. Bacteriological profile and antibiotic resistance: Comparison of findings in a burn intensive care unit, other intensive care units, and the hospital services unit of a single center. J. Burn Care Rehabil. 2005, 26, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Hossain, S.A.; Fatema, A.N.; Wahab, A.; Alam, M.M.; Islam, N.; Hossain, M.Z.; Ahsan, G.U. Age and gender-specific antibiotic resistance patterns among Bangladeshi patients with urinary tract infection caused by Escherichia coli. Heliyon 2020, 6, e04161. [Google Scholar] [CrossRef]
- Wu, L.; Zeng, T.; Deligios, M.; Milanesi, L.; Langille, M.G.I.; Zinellu, A.; Rubino, S.; Carru, C.; Kelvin, D.J. Age-Related Variation of Bacterial and Fungal Communities in Different Body Habitats across the Young, Elderly, and Centenarians in Sardinia. mSphere 2020, 5, e00558-19. [Google Scholar] [CrossRef]
- Brinkac, L.; Voorhies, A.; Gomez, A.; Nelson, K.E. The threat of antimicrobial resistance on the human microbiome. Microb. Ecol. 2017, 74, 1001–1100. [Google Scholar] [CrossRef]
- Brandl, M.; Hoffmann, A.; Willrich, N.; Reuss, A.; Reichert, F.; Walter, J.; Eckmanns, T.; Haller, S. Bugs That Can Resist Antibiotics but Not Men: Gender-Specific Differences in Notified Infections and Colonisations in Germany, 2010–2019. Microorganisms 2021, 9, 894. [Google Scholar] [CrossRef]
- Jones, N.; Mitchell, J.; Cooke, P.; Baral, S.; Arjyal, A.; Shrestha, A.; King, R. Gender and Antimicrobial Resistance: What Can We Learn from Applying a Gendered Lens to Data Analysis Using a Participatory Arts Case Study? Front. Glob. Women’s Health 2022, 3, 745862. [Google Scholar] [CrossRef]
- Aryal, B.K.; Daud, M.; Thapa, A.; Mahotra, A.; Magar, S.A.; Malla, C.K. Assesssment of health facilities for implementation of package of essential non-communicable disease in Nepal: Baseline study in Kailali and Ilam District. J. Nepal Health Res. Counc. 2018, 16, 149–155. [Google Scholar] [CrossRef]
- Shakeri, H.; Volkova, V.; Wen, X.; Deters, A.; Cull, C.; Drouillard, J.; Müller, C.; Moradijamei, B.; Jaberi-Douraki, M. Establishing Statistical Equivalence of Data from Different Sampling Approaches for Assessment of Bacterial Phenotypic Antimicrobial Resistance. Appl. Environ. Microbiol. 2018, 84, e02724-17. [Google Scholar] [CrossRef]
- Karp, B.E.; Tate, H.; Plumblee, J.R.; Dessai, U.; Whichard, J.M.; Thacker, E.L.; Hale, K.R.; Wilson, W.; Friedman, C.R.; Griffin, P.M.; et al. National Antimicrobial Resistance Monitoring System: Two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathog. Dis. 2017, 14, 545–557. [Google Scholar] [CrossRef]
- Mwansa, T.N.; Kamvuma, K.; Mulemena, J.A.; Phiri, C.N.; Chanda, W. Antibiotic susceptibility patterns of pathogens isolated from laboratory specimens at Livingstone Central Hospital in Zambia. PLoS Glob. Public Health 2022, 2, e0000623. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, S.; Zhou, Y.; Jin, S.; Ye, T.; Pan, X. Antibiotic resistance spectrum of E. coli strains from different samples and age-grouped patients: A 10-year retrospective study. BMJ Open 2023, 13, e067490. [Google Scholar] [CrossRef]
- Torres, J.; Avalos, N.; Echols, L.; Mongelluzzo, J.; Rodriguez, R.M. Low yield of blood and wound cultures in patients with skin and soft-tissue infections. Am. J. Emerg. Med. 2017, 35, 1159–1161. [Google Scholar] [CrossRef]
- WHO. Global Antimicrobial Resistance Surveillance System Manual for Early Implementation. [Internet] 2015. Available online: https://www.who.int/publications/i/item/9789241549400 (accessed on 4 March 2024).
- Baker, S.; Thomson, N.; Weill, F.-X.; Holt, K.E. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 2018, 360, 733–738. [Google Scholar] [CrossRef] [PubMed]
- CDC. Covid-19 Impact on Antimicrobial Resistance: 2022 Special Report. [Internet] 2022. Available online: https://www.cdc.gov/drugresistance/pdf/covid19-impact-report-508.pdf (accessed on 4 March 2024).
- Sivasankar, S.; Goldman, J.L.; Hoffman, M.A. Variation in antibiotic resistance patterns for children and adults treated at 166 non-affiliated US facilities using EHR data. JAC—Antimicrob. Resist. 2022, 5, dlac128. [Google Scholar] [CrossRef] [PubMed]
- Malani, A.N.; Richards, P.G.; Kapila, S.; Otto, M.H.; Czerwinski, J.; Singal, B. Clinical and economic outcomes from a community hospital’s antimicrobial stewardship program. Am. J. Infect. Control 2013, 41, 145–148. [Google Scholar] [CrossRef] [PubMed]


| Variable | Levels | Number of Specimens | Percentage of Total |
|---|---|---|---|
| Site | Jinja RRH | 2821 | 19.48% |
| DMM MUST | 2615 | 18.05% | |
| Mbarara RRH | 2384 | 16.46% | |
| Kabale RRH | 1881 | 12.99% | |
| Mbale RRH | 1487 | 10.27% | |
| Arua RRH | 1039 | 7.17% | |
| Lira RRH | 820 | 5.66% | |
| Gulu RRH | 656 | 4.53% | |
| Masaka RRH | 538 | 3.71% | |
| Soroti RRH | 243 | 1.68% | |
| Sex | Female | 8002 | 55.74% |
| Male | 6355 | 44.26% | |
| Age (Years) | 25–44 | 4902 | 33.84% |
| 5–24 | 3976 | 27.45% | |
| 0–4 | 2532 | 17.48% | |
| 45–64 | 2075 | 14.33% | |
| 65 and above | 999 | 6.90% | |
| Sample type | Urine | 3279 | 25.29% |
| Blood | 3245 | 25.03% | |
| Pus Swab | 2774 | 21.39% | |
| Urogenital swabs | 1781 | 13.73% | |
| Cerebral Spinal Fluid | 881 | 6.79% | |
| Sputum | 564 | 4.35% | |
| Stool | 407 | 3.14% | |
| Tracheal aspirate | 36 | 0.28% | |
| Quarter | October–December 2020 | 1936 | 13.37% |
| January–March 2021 | 2159 | 14.91% | |
| April–June 2021 | 2093 | 14.45% | |
| July–September 2021 | 1881 | 12.99% | |
| October–December 2021 | 1366 | 9.43% | |
| January–March 2022 | 1120 | 7.73% | |
| April–June 2022 | 667 | 4.61% | |
| July–September 2022 | 677 | 4.67% | |
| October–December 2022 | 852 | 5.88% | |
| January–March 2023 | 1733 | 11.96% | |
| Department | Outpatient department | 5282 | 45.31% |
| Paediatric ward | 2141 | 18.37% | |
| Medical ward | 1365 | 11.71% | |
| Surgical ward | 885 | 7.59% | |
| Gynaecology ward | 547 | 4.69% | |
| Emergency ward | 537 | 4.61% | |
| Orthopaedic department | 266 | 2.28% | |
| Maternity ward | 168 | 1.44% | |
| Private ward | 134 | 1.15% | |
| Acute Care Unit | 133 | 1.14% | |
| Intensive care unit | 106 | 0.91% | |
| HIV clinic | 94 | 0.81% |
| Antibiotic | Gram-Negative Isolates | Gram-Positive Isolates | |||||
|---|---|---|---|---|---|---|---|
| Enterobacterales | Non Enterobacterales | ||||||
| E. coli n (%S) | Klebsiella spp. n (%S) | Others n (%S) | Acinetobacter spp. n (%S) | Pseudomonas spp. n (%S) | Staphylococcus aureus n (%S) | Enterococcus spp. n (%S) | |
| Ampicillin | 607 (8%) | 79 (22%) | 70 (66%) | ||||
| Amoxicillin/Clavulanic acid | 463 (56%) | 11 (82%) | |||||
| Ciprofloxacin | 650 (36%) | 299 (55%) | 220 (58%) | 69 (54%) | 73 (60%) | 799 (51%) | 86 (51%) |
| Chloramphenicol | 411 (67%) | 179 (56%) | 124 (39%) | 631 (68%) | 49 (59%) | ||
| Ceftriaxone | 375 (28%) | 226 (38%) | 155 (42%) | 31 (16%) | |||
| Meropenem | 172 (81%) | 86 (90%) | 77 (75%) | 20 (55%) | 19 (63%) | ||
| Imipenem | 629 (89%) | 365 (90%) | 201 (77%) | 59 (66%) | 94 (87%) | ||
| Trimethoprim | 466 (15%) | 275 (21%) | 167 (36%) | 40 (20%) | 597 (32%) | ||
| Gentamicin | 597 (57%) | 278 (54%) | 192 (71%) | 57 (53%) | 60 (65%) | 718 (66%) | |
| Erythromycin | 748 (34%) | ||||||
| Vancomycin | 86 (72%) | ||||||
| Cefoxitin (MRSA Screen) | 172 (44%) | ||||||
| Antibiotic | Emergency n (%S) | Gynaecology n (%S) | HIV Clinic n (%S) | Maternity n (%S) | Medical n (%S) | OPD n (%S) | Paediatrics n (%S) | Specialized n (%S) | Surgical n (%S) | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | 38 (18%) | 83 (14%) | 12 * (33%) | 27 * (7%) | 150 (13%) | 608 (17%) | 84 (10%) | 24 * (4%) | 230 (7%) | 0 |
| Amoxicillin/Clavulanic acid | 23 * (35%) | 71 (45%) | 9 * (56%) | 16 * (50%) | 121 (42%) | 434 (54%) | 70 (41%) | 9 * (44%) | 188 (47%) | 0.007 |
| Ciprofloxacin | 80 (46%) | 157 (44%) | 17 * (33%) | 47 (36%) | 251 (46%) | 886 (48%) | 195 (56%) | 35 (49%) | 416 (44%) | 0.888 |
| Chloramphenicol | 32 (56%) | 93 (61%) | 11 * (82%) | 42 (55%) | 137 (66%) | 622 (60%) | 139 (69%) | 24 * (50%) | 318 (58%) | 0.109 |
| Ceftriaxone | 18 * (17%) | 58 (21%) | 7 * (43%) | 8 * (25%) | 115 (27%) | 315 (57%) | 76 (26%) | 14 * (21%) | 196 (22%) | 0 |
| Meropenem | 8 * (87%) | 27 * (74%) | 5 * (40%) | 55 (89%) | 161 (86%) | 19 * (79%) | 2 * (50%) | 67 (67%) | 0.136 | |
| Imipenem | 51 (90%) | 96 (77%) | 12 * (92%) | 27 * (89%) | 194 (89%) | 627 (90%) | 112 (85%) | 32 (75%) | 249 (84%) | 0.025 |
| Trimethoprim/Sulfamethoxazole | 31 (6%) | 96 (19%) | 13 * (8%) | 48 (21%) | 173 (20%) | 694 (24%) | 161 (25%) | 17 * (24%) | 240 (21%) | 0 |
| Gentamicin | 61 (70%) | 126 (44%) | 18 * (67%) | 50 (56%) | 193 (64%) | 767 (65%) | 153 (51%) | 23 * (61%) | 400 (55%) | 0 |
| Erythromycin | 24 * (33%) | 57 (23%) | 12 * (33%) | 24 * (25%) | 90 (41%) | 527 (26%) | 130 (32%) | 11 * (27%) | 157 (45%) | 0 |
| Vancomycin | 5 * (20%) | 26 * (46%) | 1 * (0%) | 3 * (67%) | 48 (42%) | 249 (36%) | 28 * (61%) | 6 * (67%) | 24 * (21%) | 0.64 |
| Oxacillin/Methicillin | 10 * (50%) | 9 * (44%) | 1 * (0%) | 2 * (50%) | 17 * (53%) | 82 (44%) | 38 (47%) | 1 * (0%) | 37 (59%) | 0 |
| Antibiotic Name | Age Groups | Susceptibility | OR [95% CI] | p-Values |
|---|---|---|---|---|
| Ampicillin | 0–4 | 173 (13%) | 0.86 [0.468–1.588] | 0.634 |
| 5–9 | 41 (21%) | 0.45 [0.191–1.043] | 0.063 | |
| 10–14 | 40 (22%) | 0.43 [0.185–1.013] | 0.054 | |
| 15–25 | 294 (18%) | 0.57 [0.343–0.952] | 0.032 | |
| 25–44 | 556 (12%) | 0.86 [0.528–1.393] | 0.536 | |
| 45–64 | 332 (14%) | 0.74 [0.444–1.246] | 0.26 | |
| 65 and above | 224 (11%) | Ref | ||
| Amoxiclav | 0–4 | 117 (47%) | 1.24 [0.773–1.985] | 0.374 |
| 5–9 | 34 (58%) | 0.77 [0.365–1.622] | 0.491 | |
| 10–14 | 31 (45%) | 1.33 [0.619–2.878] | 0.462 | |
| 15–25 | 205 (55%) | 0.88 [0.583–1.319] | 0.529 | |
| 25–44 | 386 (46%) | 1.24 [0.867–1.786] | 0.235 | |
| 45–64 | 240 (46%) | 1.28 [0.861–1.893] | 0.224 | |
| 65 and above | 170 (52%) | Ref | ||
| Ciprofloxacin | 0–4 | 377 (58%) | 0.73 [0.532–0.993] | 0.045 |
| 5–9 | 67 (62%) | 0.6 [0.346–1.038] | 0.068 | |
| 10–14 | 91 (74%) | 0.34 [0.201–0.578] | 0.000 | |
| 15–25 | 567 (47%) | 1.12 [0.837–1.489] | 0.456 | |
| 25–44 | 964 (41%) | 1.4 [1.072–1.835] | 0.014 | |
| 45–64 | 505 (45%) | 1.19 [0.883–1.591] | 0.257 | |
| 65 and above | 275 (50%) | Ref | ||
| Chloramphenicol | 0–4 | 253 (64%) | 1.15 [0.757–1.733] | 0.520 |
| 5–9 | 55 (69%) | 0.94 [0.49–1.822] | 0.865 | |
| 10–14 | 50 (68%) | 0.99 [0.505–1.954] | 0.985 | |
| 15–25 | 397 (65%) | 1.09 [0.74–1.598] | 0.668 | |
| 25–44 | 614 (56%) | 1.66 [1.155–2.377] | 0.006 | |
| 45–64 | 332 (57%) | 1.54 [1.042–2.274] | 0.030 | |
| 65 and above | 168 (67%) | Ref | ||
| Ceftriaxone | 0–4 | 105 (27%) | 1.64 [0.936–2.881] | 0.084 |
| 5–9 | 34 (38%) | 1.01 [0.463–2.213] | 0.976 | |
| 10–14 | 29 (58%) | 0.44 [0.194–1.008] | 0.052 | |
| 15–25 | 179 (34%) | 1.21 [0.752–1.955] | 0.430 | |
| 25–44 | 337 (38%) | 1 [0.652–1.527] | 0.992 | |
| 45–64 | 216 (31%) | 1.36 [0.857–2.169] | 0.190 | |
| 65 and above | 122 (38%) | Ref | ||
| Meropenem | 0–4 | 53 (77%) | 2.2 [0.825–5.842] | 0.115 |
| 5–9 | 15 (86%) | 1.15 [0.219–6.077] | 0.866 | |
| 10–14 | 10 (80%) | 1.88 [0.337–10.431] | 0.473 | |
| 15–25 | 74 (77%) | 2.24 [0.896–5.586] | 0.085 | |
| 25–44 | 159 (80%) | 1.82 [0.788–4.189] | 0.161 | |
| 45–64 | 90 (82%) | 1.62 [0.65–4.047] | 0.300 | |
| 65 and above | 68 (88%) | Ref | ||
| Imipenem | 0–4 | 197 (82%) | 1.73 [0.991–3.011] | 0.054 |
| 5–9 | 43 (86%) | 1.34 [0.516–3.497] | 0.546 | |
| 10–14 | 61 (85%) | 1.43 [0.631–3.255] | 0.390 | |
| 15–25 | 314 (85%) | 1.35 [0.8–2.277] | 0.262 | |
| 25–44 | 596 (85%) | 1.36 [0.845–2.184] | 0.206 | |
| 45–64 | 364 (84%) | 1.51 [0.91–2.49] | 0.111 | |
| 65 and above | 232 (89%) | Ref | ||
| Trimethoprim Sulphamethoxazole | 0–4 | 267 (25%) | 0.79 [0.512–1.223] | 0.292 |
| 5–9 | 63 (31%) | 0.58 [0.31–1.09] | 0.091 | |
| 10–14 | 69 (34%) | 0.51 [0.279–0.923] | 0.026 | |
| 15–25 | 360 (22%) | 0.93 [0.613–1.415] | 0.739 | |
| 25–44 | 670 (22%) | 0.94 [0.639–1.375] | 0.741 | |
| 45–64 | 331 (25%) | 0.81 [0.532–1.228] | 0.318 | |
| 65 and above | 202 (21%) | Ref | ||
| Gentamicin | 0–4 | 307 (51%) | 1.7 [1.199–2.402] | 0.003 |
| 5–9 | 57 (73%) | 0.63 [0.332–1.211] | 0.168 | |
| 10–14 | 70 (80%) | 0.44 [0.233–0.845] | 0.013 | |
| 15–25 | 494 (60%) | 1.15 [0.833–1.584] | 0.398 | |
| 25–44 | 832 (58%) | 1.27 [0.942–1.715] | 0.116 | |
| 45–64 | 420 (59%) | 1.22 [0.878–1.696] | 0.237 | |
| 65 and above | 236 (63%) | Ref | ||
| Erythromycin | 0–4 | 215 (31%) | 0.99 [0.577–1.695] | 0.969 |
| 5–9 | 49 (44%) | 0.56 [0.272–1.159] | 0.118 | |
| 10–14 | 60 (50%) | 0.46 [0.232–0.904] | 0.024 | |
| 15–25 | 300 (35%) | 0.85 [0.509–1.42] | 0.535 | |
| 25–44 | 477 (28%) | 1.14 [0.691–1.866] | 0.616 | |
| 45–64 | 196 (30%) | 1.04 [0.6–1.793] | 0.896 | |
| 65 and above | 86 (31%) | Ref | ||
| Vancomycin | 0–4 | 59 (44%) | 0.71 [0.33–1.547] | 0.393 |
| 5–9 | 17 (47%) | 0.63 [0.208–1.927] | 0.421 | |
| 10–14 | 15 (40%) | 0.84 [0.258–2.755] | 0.778 | |
| 15–25 | 72 (37%) | 0.94 [0.443–1.983] | 0.866 | |
| 25–44 | 150 (40%) | 0.82 [0.423–1.593] | 0.559 | |
| 45–64 | 97 (38%) | 0.91 [0.449–1.852] | 0.799 | |
| 65 and above | 50 (36%) | Ref | ||
| Oxacillin/Methicillin | 0–4 | 55 (38%) | 0.4 [0.102–1.604] | 0.198 |
| 5–9 | 9 (55%) | 0.2 [0.032–1.24] | 0.084 | |
| 10–14 | 18 (77%) | 0.07 [0.013–0.385] | 0.002 | |
| 15–25 | 52 (42%) | 0.34 [0.086–1.355] | 0.126 | |
| 25–44 | 69 (46%) | 0.29 [0.075–1.116] | 0.072 | |
| 45–64 | 35 (45%) | 0.3 [0.071–1.24] | 0.096 | |
| 65 and above | 15 (20%) | Ref |
| Antibiotic | Gram-Negatives | Gram-Positives | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acinetobacter spp. | Escherichia coli | Klebsiella spp. | Staphylococcus aureus | Enterococcus spp. | ||||||||||||||||
| Blood | Urine | Pus | p-Value | Blood | Urine | Pus | p-Values | Blood | Urine | Pus | p-Values | Blood | Urine | Pus | p-Value | Blood | Urine | Pus | p-Value | |
| n (%S) | n (%S) | n (%S) | n (%S) | n (%S) | n (%S) | n (%S) | n (%S) | n (%S) | n (%S) | n (%S) | n (%S) | n (%R) | n (%S) | n (%S) | ||||||
| Ampicillin | NA | NA | NA | 20 (20%) | 283 (9%) | 194 (5%) | 0.088 | NA | NA | NA | NA | NA | NA | NA | 14 (36%) | 27 (67%) | 8 (87%) | 0.046 | ||
| Amoxicillin/Clavulanic acid | NA | NA | NA | 21 (57%) | 216 (53%) | 147 (56%) | 0.705 | 22 (36%) | 55 (44%) | 84 (42%) | NA | NA | NA | NA | NA | NA | ||||
| Ciprofloxacin | 2 (100%) | 5 (40%) | 44 (57%) | 0.047 | 23 (57%) | 256 (39%) | 246 (28%) | 0.005 | 29 (41%) | 50 (62%) | 126 (49%) | 0.121 | 82 (62%) | 153 (37%) | 407 (62%) | 0.000 | 13 (15%) | 39 (64%) | 13 (31%) | 0.004 |
| Chloramphenicol | NA | NA | NA | 12 (33%) | 133 (72%) | 150 (75%) | 0.021 | 17 (41%) | 30 (77%) | 89 (39%) | 0.008 | 60 (77%) | 128 (58%) | 309 (75%) | 0.005 | 12 (67%) | 17 (59%) | 10 (70%) | 0.689 | |
| Ceftriaxone | 16 (44%) | 149 (34%) | 133 (14%) | 0.002 | 27 (19%) | 45 (56%) | 75 (28%) | 0.006 | NA | NA | NA | NA | NA | NA | ||||||
| Meropenem | 4 (50%) | 13 (54%) | 6 (83%) | 62 (85%) | 63 (79%) | 0.902 | 5 (80%) | 16 (94%) | 34 (88%) | 0.636 | NA | NA | NA | NA | NA | NA | ||||
| Imipenem | 5 (80%) | 3 (100%) | 34 (56%) | 0.765 | 26 (81%) | 314 (89%) | 200 (86%) | 0.302 | 28 (100%) | 78 (91%) | 121 (83%) | 0.088 | NA | NA | NA | NA | NA | NA | ||
| Trimethoprim/Sulfamethoxazole | 3 (33%) | 1 (100%) | 23 (13%) | 0.144 | 15 (27%) | 201 (10%) | 162 (14%) | 0.33 | 27 (26%) | 65 (22%) | 84 (15%) | 0.473 | 58 (28%) | 135 (21%) | 282 (42%) | 0.000 | NA | NA | NA | |
| Gentamicin | 2 (100%) | 5 (40%) | 35 (54%) | 0.47 | 18 (61%) | 230 (58%) | 223 (49%) | 0.171 | 31 (39%) | 45 (60%) | 109 (42%) | 0.049 | 69 (58%) | 142 (61%) | 350 (71%) | 0.058 | NA | NA | NA | |
| Piperacillin | 1 (100%) | 1 (0%) | 2 (50%) | NA | NA | NA | NA | NA | NA | |||||||||||
| Erythromycin | NA | NA | NA | NA | NA | NA | NA | NA | NA | 70 (46%) | 179 (23%) | 352 (41%) | 0.000 | NA | NA | NA | ||||
| Vancomycin | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 16 (81%) | 38 (74%) | 12 (58%) | 0.440 | |||||
| Oxacillin/ Methicillin | NA | NA | NA | NA | NA | NA | NA | NA | 17 (47%) | 27 (26%) | 99 (51%) | 0.123 | NA | NA | NA | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayito, J.; Kibombo, D.; Olaro, C.; Nabadda, S.; Guma, C.; Nabukenya, I.; Busuge, A.; Dhikusooka, F.; Andema, A.; Mukobi, P.; et al. Characterization of Antibiotic Resistance in Select Tertiary Hospitals in Uganda: An Evaluation of 2020 to 2023 Routine Surveillance Data. Trop. Med. Infect. Dis. 2024, 9, 77. https://doi.org/10.3390/tropicalmed9040077
Mayito J, Kibombo D, Olaro C, Nabadda S, Guma C, Nabukenya I, Busuge A, Dhikusooka F, Andema A, Mukobi P, et al. Characterization of Antibiotic Resistance in Select Tertiary Hospitals in Uganda: An Evaluation of 2020 to 2023 Routine Surveillance Data. Tropical Medicine and Infectious Disease. 2024; 9(4):77. https://doi.org/10.3390/tropicalmed9040077
Chicago/Turabian StyleMayito, Jonathan, Daniel Kibombo, Charles Olaro, Susan Nabadda, Consolata Guma, Immaculate Nabukenya, Andrew Busuge, Flavia Dhikusooka, Alex Andema, Peter Mukobi, and et al. 2024. "Characterization of Antibiotic Resistance in Select Tertiary Hospitals in Uganda: An Evaluation of 2020 to 2023 Routine Surveillance Data" Tropical Medicine and Infectious Disease 9, no. 4: 77. https://doi.org/10.3390/tropicalmed9040077
APA StyleMayito, J., Kibombo, D., Olaro, C., Nabadda, S., Guma, C., Nabukenya, I., Busuge, A., Dhikusooka, F., Andema, A., Mukobi, P., Onyachi, N., Watmon, B., Obbo, S., Yayi, A., Elima, J., Barigye, C., Nyeko, F. J., Mugerwa, I., Sekamatte, M., ... Kajumbula, H. (2024). Characterization of Antibiotic Resistance in Select Tertiary Hospitals in Uganda: An Evaluation of 2020 to 2023 Routine Surveillance Data. Tropical Medicine and Infectious Disease, 9(4), 77. https://doi.org/10.3390/tropicalmed9040077





