Structures and Functional Diversities of ASFV Proteins
Abstract
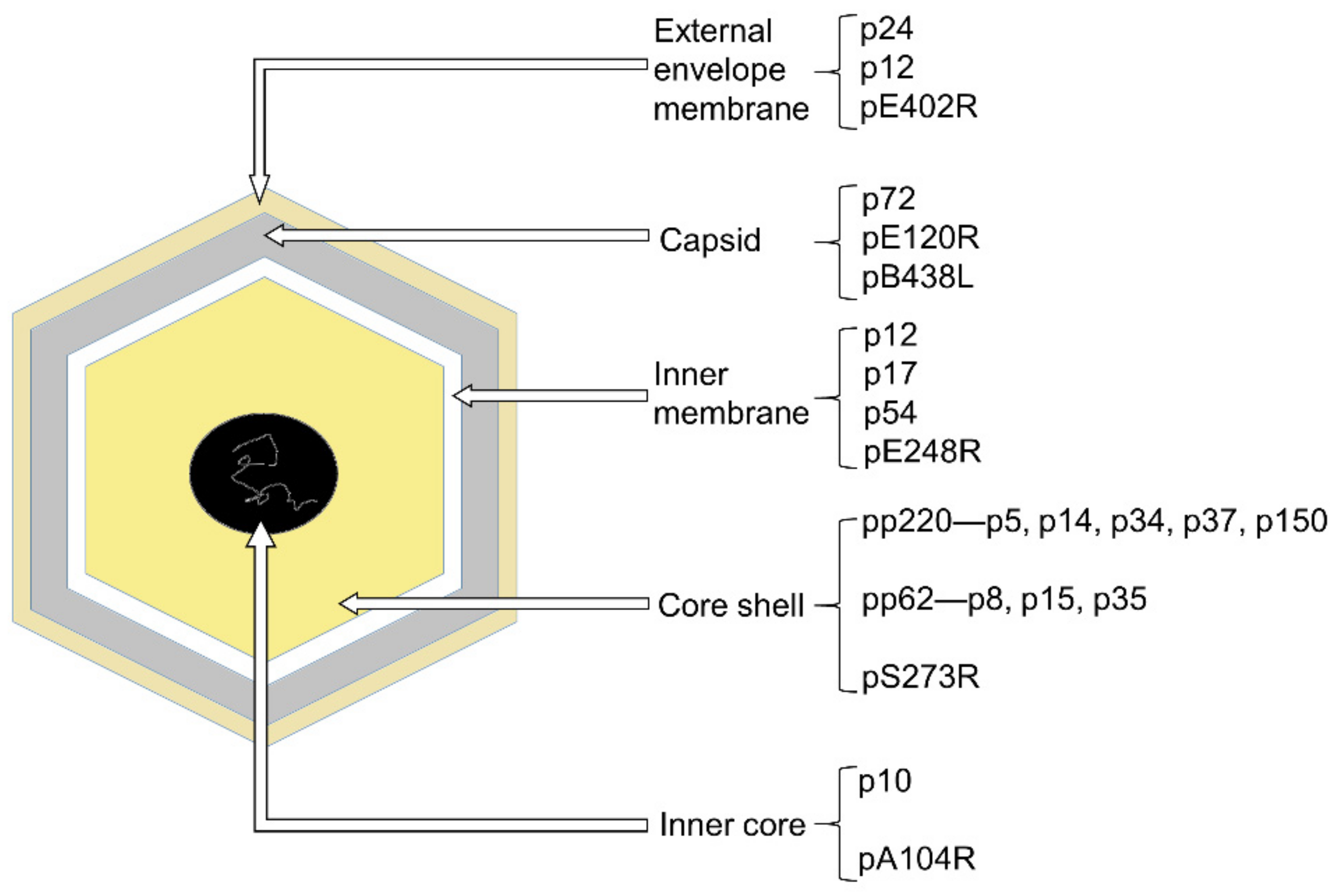
:1. Introduction
2. A Few Structures, Functions, and Mechanisms of ASFV Proteins Have Been Revealed
2.1. AsfvAP, AsfvPolX, and AsfvLIG Are Essential in ASFV Genome Repair and Variation
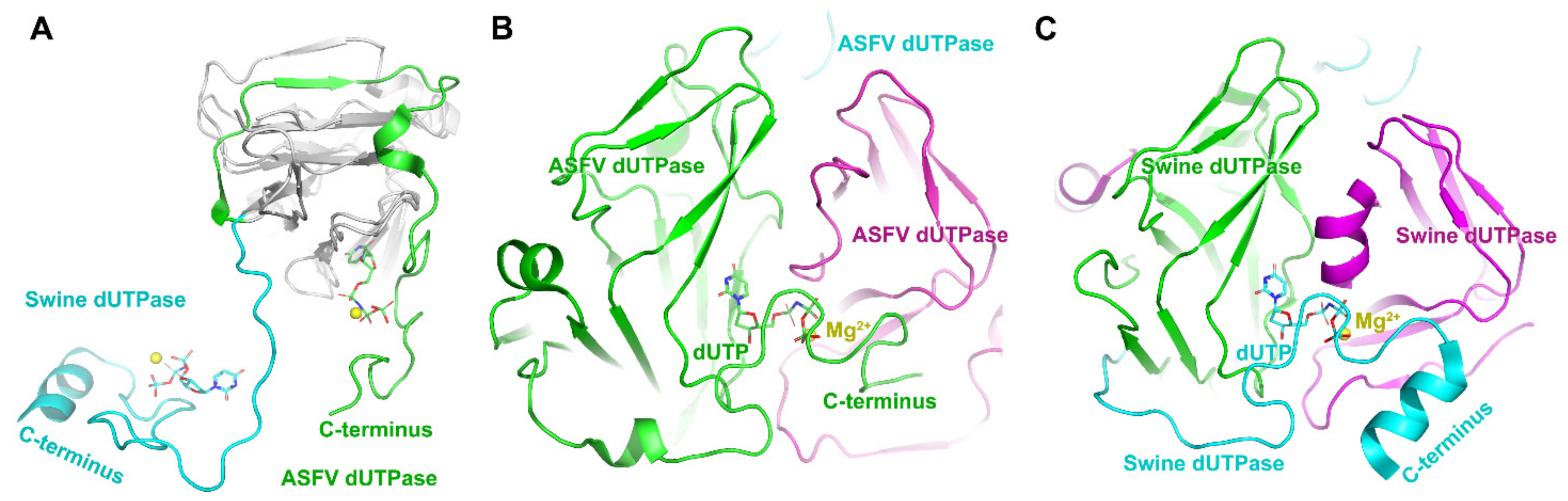
2.2. ASFV dUTPase and Guanylyltransferase
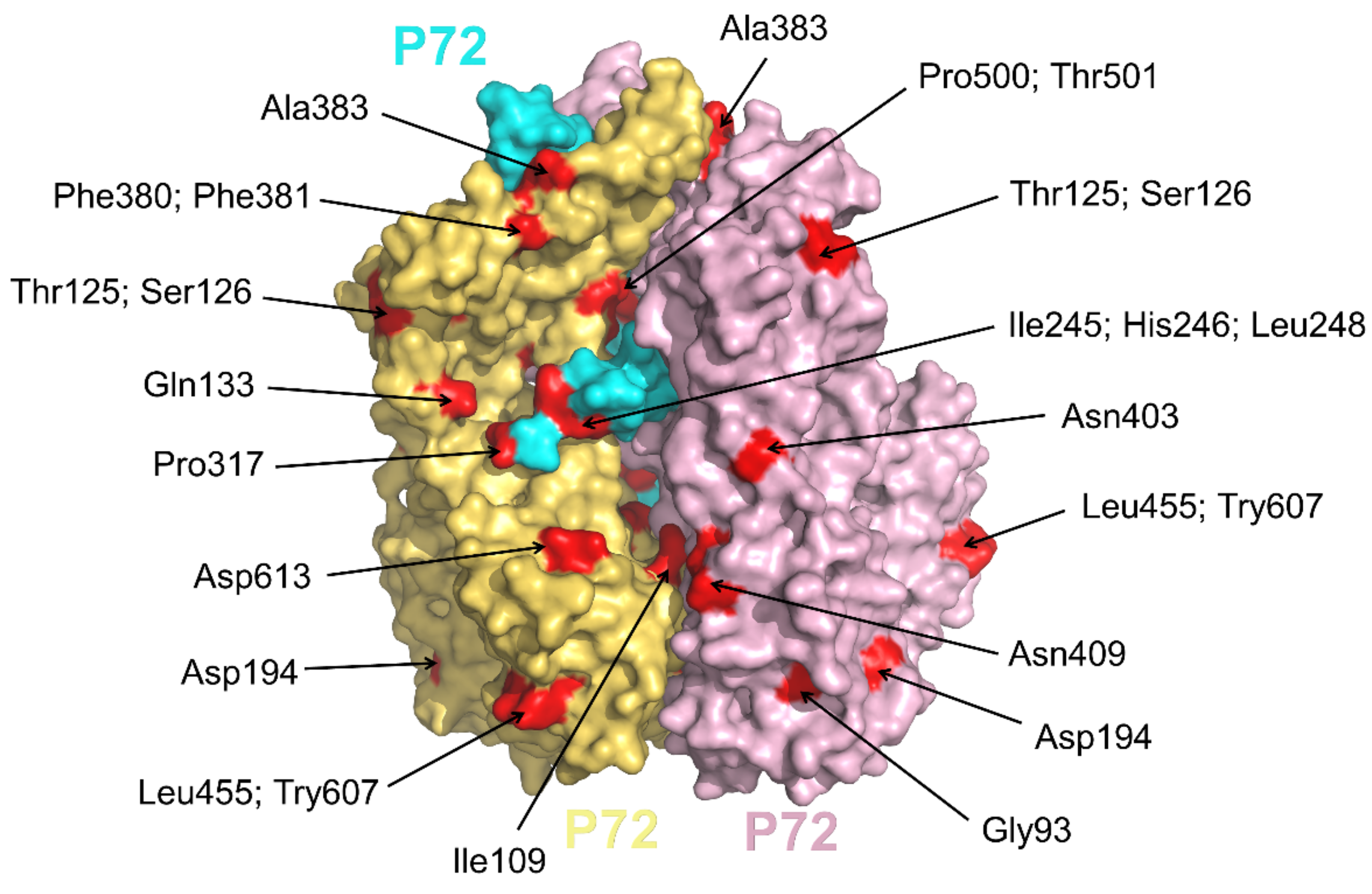
2.3. The p72 Is a Major Component of Capsid
2.4. Structural Proteins P15 and p35 Are Cleaved from pp62 by Protease pS273R
2.5. Sulfhydryl Oxidase pB119L and Thioredoxin pA151R
2.6. pA104R, an Important DNA-Binding Protein in Replication
2.7. A179L, a Potent Protein Inhibitor of Apoptosis
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, F.; Shi, Y.; Yang, M.; Guo, Y.; Shen, Z.; Li, M.; Chen, Y.; Liang, R.; Yang, Y.; Chen, H.; et al. The structural basis of African swine fever virus core shell protein p15 binding to DNA. FASEB J. 2021, 35, e21350. [Google Scholar] [CrossRef] [PubMed]
- Xian, Y.; Xiao, C. The Structure of ASFV Advances the Fight against the Disease. Trends Biochem. Sci. 2020, 45, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Penrith, M.L.; Vosloo, W. Review of African swine fever: Transmission, spread and control. J. S. Afr. Vet. Assoc. 2009, 80, 58–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, L.K.; Sun, H.; Roberts, H. African swine fever. Antiviral Res. 2019, 165, 34–41. [Google Scholar] [CrossRef]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andrés, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92, e01293-18. [Google Scholar] [CrossRef] [Green Version]
- Dixon, L.K.; Chapman, D.A.G.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, D.; Wang, J.; Zhang, Y.; Wang, M.; Gao, Y.; Li, F.; Wang, J.; Bu, Z.; Rao, Z.; et al. Architecture of African swine fever virus and implications for viral assembly. Science 2019, 366, 640–644. [Google Scholar] [CrossRef]
- Liu, S.; Luo, Y.; Wang, Y.; Li, S.; Zhao, Z.; Bi, Y.; Sun, J.; Peng, R.; Song, H.; Zhu, D.; et al. Cryo-EM Structure of the African Swine Fever Virus. Cell Host Microbe. 2019, 26, 836–843.e833. [Google Scholar] [CrossRef]
- Breese, S.S., Jr.; DeBoer, C.J. Electron microscope observations of African swine fever virus in tissue culture cells. Virology 1966, 28, 420–428. [Google Scholar] [CrossRef]
- Hawes, P.C.; Netherton, C.L.; Wileman, T.E.; Monaghan, P. The envelope of intracellular African swine fever virus is composed of a single lipid bilayer. J. Virol. 2008, 82, 7905–7912. [Google Scholar] [CrossRef] [Green Version]
- Andrés, G.; García-Escudero, R.; Simón-Mateo, C.; Viñuela, E. African Swine Fever Virus Is Enveloped by a Two-Membraned Collapsed Cisterna Derived from the Endoplasmic Reticulum. J. Virol. 1998, 72, 8988–9001. [Google Scholar] [CrossRef]
- Andres, G.; Simon-Mateo, C.; Vinuela, E. Assembly of African swine fever virus: Role of polyprotein pp220. J. Virol. 1997, 71, 2331–2341. [Google Scholar] [CrossRef] [Green Version]
- Reis, A.L.; Netherton, C.; Dixon, L.K. Unraveling the Armor of a Killer: Evasion of Host Defenses by African Swine Fever Virus. J. Virol. 2017, 91, e02338-16. [Google Scholar] [CrossRef] [Green Version]
- Takamatsu, H.-H.; Denyer, M.S.; Lacasta, A.; Stirling, C.M.A.; Argilaguet, J.M.; Netherton, C.L.; Oura, C.A.L.; Martins, C.; Rodríguez, F. Cellular immunity in ASFV responses. Virus Res. 2013, 173, 110–121. [Google Scholar] [CrossRef]
- Thomson, G.R.; Gainaru, M.D.; Van Dellen, A.F. Experimental infection of warthos (Phacochoerus aethiopicus) with African swine fever virus. Onderstepoort J. Vet. Res. 1980, 47, 19–22. [Google Scholar]
- Correia, S.; Ventura, S.; Parkhouse, R.M. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 2013, 173, 87–100. [Google Scholar] [CrossRef]
- Ekue, N.F.; Wilkinson, P.J. Absence of Ornithodoros moubata, the vector of african swine fever virus, from the main pig producing area of Cameroon. Trop. Anim. Health Prod. 1990, 22, 127–131. [Google Scholar] [CrossRef]
- Wilkinson, P.J.; Donaldson, A.I.; Greig, A.; Bruce, W. Transmission studies with African swine fever virus: Infections of pigs by airborne virus. J. Comp. Pathol. 1977, 87, 487–495. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, X.; Huang, Q.; Shao, Z.; Gao, Y.; Li, Y.; Yang, C.; Liu, H.; Li, J.; Wang, Q.; et al. A unique DNA-binding mode of African swine fever virus AP endonuclease. Cell Discov. 2020, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Showalter, A.K.; Byeon, I.J.; Su, M.I.; Tsai, M.D. Solution structure of a viral DNA polymerase X and evidence for a mutagenic function. Nat. Struct. 2001, 8, 942–946. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, J.; Liu, H.; Gao, Y.; Li, X.; Zheng, L.; Cui, R.; Yao, Q.; Rong, L.; Li, J.; et al. Unique 5’-P recognition and basis for dG:dGTP misincorporation of ASFV DNA polymerase X. PLoS Biol. 2017, 15, e1002599. [Google Scholar] [CrossRef]
- Yanez, R.J.; Rodriguez, J.M.; Nogal, M.L.; Yuste, L.; Enriquez, C.; Rodriguez, J.F.; Vinuela, E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 1995, 208, 249–278. [Google Scholar] [CrossRef] [Green Version]
- Akaike, T. Role of free radicals in viral pathogenesis and mutation. Annu. Rev. Virol. 2001, 11, 87–101. [Google Scholar] [CrossRef]
- Rojo, G.; Garcia-Beato, R.; Vinuela, E.; Salas, M.L.; Salas, J. Replication of African swine fever virus DNA in infected cells. Virology 1999, 257, 524–536. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Liu, H.; Yang, C.; Gao, Y.; Yu, X.; Chen, X.; Cui, R.; Zheng, L.; Li, S.; Li, X.; et al. Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues. Nat. Commun. 2019, 10, 387. [Google Scholar] [CrossRef] [Green Version]
- Pascal, J.M.; O’Brien, P.J.; Tomkinson, A.E.; Ellenberger, T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 2004, 432, 473–478. [Google Scholar] [CrossRef]
- De Ioannes, P.; Malu, S.; Cortes, P.; Aggarwal, A.K. Structural basis of DNA ligase IV-Artemis interaction in nonhomologous end-joining. Cell Rep. 2012, 2, 1505–1512. [Google Scholar] [CrossRef] [Green Version]
- Lamarche, B.J.; Kumar, S.; Tsai, M.-D. ASFV DNA polymerse X is extremely error-prone under diverse assay conditions and within multiple DNA sequence contexts. Biochemistry 2006, 45, 14826–14833. [Google Scholar] [CrossRef] [Green Version]
- García-Escudero, R.; García-Díaz, M.; Salas, M.a.L.; Blanco, L.; Salas, J. DNA Polymerase X of African Swine Fever Virus: Insertion Fidelity on Gapped DNA substrates and AP lyase Activity Support a Role in Base Excision Repair of Viral DNA. J. Mol. Biol. 2003, 326, 1403–1412. [Google Scholar] [CrossRef]
- Lapenna, A.; Stefan, A.; Hochkoeppler, A. ASFV DNA polymerase extends recessed DNAs with catalytic efficiencies outperforming those exerted on gapped DNA substrates. Biochem. Biophys. Res. Commun. 2021, 534, 526–532. [Google Scholar] [CrossRef]
- Liang, R.; Wang, G.; Zhang, D.; Ye, G.; Li, M.; Shi, Y.; Shi, J.; Chen, H.; Peng, G. Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development. J. Biol. Chem. 2021, 296, 100015. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Larsson, G.; Persson, R.; Cedergren-Zeppezauer, E. Atomic resolution structure of Escherichia coli dUTPase determined ab initio. Acta Crystallogr. 2001, 57, 767–774. [Google Scholar] [CrossRef] [Green Version]
- Vértessy, B.G.; Tóth, J. Keeping Uracil Out of DNA: Physiological Role, Structure and Catalytic Mechanism of dUTPases. Acc. Chem. Res. 2009, 42, 97–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveros, M.; García-Escudero, R.; Alejo, A.; Viñuela, E.; Salas María, L.; Salas, J. African Swine Fever Virus dUTPase Is a Highly Specific Enzyme Required for Efficient Replication in Swine Macrophages. J. Virol. 1999, 73, 8934–8943. [Google Scholar] [CrossRef] [Green Version]
- Shuman, S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 2001, 66, 1–40. [Google Scholar] [CrossRef]
- Decroly, E.; Ferron, F.; Lescar, J.; Canard, B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2011, 10, 51–65. [Google Scholar] [CrossRef]
- Pena, L.; Yanez, R.J.; Revilla, Y.; Vinuela, E.; Salas, M.L. African swine fever virus guanylyltransferase. Virology 1993, 193, 319–328. [Google Scholar] [CrossRef]
- Du, X.; Gao, Z.Q.; Geng, Z.; Dong, Y.H.; Zhang, H. Structure and Biochemical Characteristic of the Methyltransferase (MTase) Domain of RNA Capping Enzyme from African Swine Fever Virus. J. Virol. 2020, 98, e02029-20. [Google Scholar] [CrossRef]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H.; et al. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerging Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Ma, B.; Qian, N.; Zhang, F.; Tan, X.; Lei, J.; Xiang, Y. Structure of the African swine fever virus major capsid protein p72. Cell Res. 2019, 29, 953–955. [Google Scholar] [CrossRef] [Green Version]
- Hakizimana, J.N.; Nyabongo, L.; Ntirandekura, J.B.; Yona, C.; Ntakirutimana, D.; Kamana, O.; Nauwynck, H.; Misinzo, G. Genetic Analysis of African Swine Fever Virus From the 2018 Outbreak in South-Eastern Burundi. Front. Vet. Sci. 2020, 7, 578474. [Google Scholar] [CrossRef]
- Simon-Mateo, C.; Andres, G.; Almazan, F.; Vinuela, E. Proteolytic processing in African swine fever virus: Evidence for a new structural polyprotein, pp62. J. Virol. 1997, 71, 5799–5804. [Google Scholar] [CrossRef] [Green Version]
- Andrés, G.; Alejo, A.; Salas, J.; Salas, M.L. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J. Virol. 2002, 76, 12473–12482. [Google Scholar] [CrossRef] [Green Version]
- Suarez, C.; Salas, M.L.; Rodriguez, J.M. African swine fever virus polyprotein pp62 is essential for viral core development. J. Virol. 2010, 84, 176–187. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Fu, D.; Zhang, G.; Zhao, D.; Li, M.; Geng, X.; Sun, D.; Wang, Y.; Chen, C.; Jiao, P.; et al. Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly. Protein Cell 2020, 11, 600–605. [Google Scholar] [CrossRef]
- Li, G.; Liu, X.; Yang, M.; Zhang, G.; Wang, Z.; Guo, K.; Gao, Y.; Jiao, P.; Sun, J.; Chen, C.; et al. Crystal Structure of African Swine Fever Virus pS273R Protease and Implications for Inhibitor Design. J. Virol. 2020, 94, e02125-19. [Google Scholar] [CrossRef]
- Andres, G.; Alejo, A.; Simon-Mateo, C.; Salas, M.L. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 2001, 276, 780–787. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, I.; Redrejo-Rodriguez, M.; Rodriguez, J.M.; Alejo, A.; Salas, J.; Salas, M.L. African swine fever virus pB119L protein is a flavin adenine dinucleotide-linked sulfhydryl oxidase. J. Virol. 2006, 80, 3157–3166. [Google Scholar] [CrossRef] [Green Version]
- Hakim, M.; Fass, D. Dimer interface migration in a viral sulfhydryl oxidase. J. Mol. Biol. 2009, 391, 758–768. [Google Scholar] [CrossRef]
- Keita, D.; Heath, L.; Albina, E. Control of African swine fever virus replication by small interfering RNA targeting the A151R and VP72 genes. Antivir. Ther. 2010, 15, 727–736. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.W.; Niu, D.; Liu, K.; Wang, Q.; Ma, L.; Chen, C.C.; Zhang, L.; Liu, W.; Zhou, S.; Min, J.; et al. Structure basis of non-structural protein pA151R from African Swine Fever Virus. Biochem. Biophys. Res. Commun. 2020, 532, 108–113. [Google Scholar] [CrossRef]
- Jeng, M.F.; Campbell, A.P.; Begley, T.; Holmgren, A.; Case, D.A.; Wright, P.E.; Dyson, H.J. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure 1994, 2, 853–868. [Google Scholar] [CrossRef] [Green Version]
- Ellgaard, L.; Ruddock, L.W. The human protein disulphide isomerase family: Substrate interactions and functional properties. EMBO Rep. 2005, 6, 28–32. [Google Scholar] [CrossRef]
- Frouco, G.; Freitas, F.B.; Coelho, J.; Leitão, A.; Martins, C.; Ferreira, F. DNA-binding properties of african swine fever virus pA104R, a histone-like protein involved in viral replication and transcription. J. Virol. 2017, 91, e02498-16. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Sun, Y.; Chai, Y.; Li, S.; Li, S.; Wang, L.; Su, J.; Yu, S.; Yan, J.; Gao, F.; et al. The structural basis of African swine fever virus pA104R binding to DNA and its inhibition by stilbene derivatives. Proc. Natl. Acad. Sci. USA 2020, 117, 11000–11009. [Google Scholar] [CrossRef]
- Revilla, Y.; Cebrian, A.; Baixeras, E.; Martinez, C.; Vinuela, E.; Salas, M.L. Inhibition of apoptosis by the African swine fever virus Bcl-2 homologue: Role of the BH1 domain. Virology 1997, 228, 400–404. [Google Scholar] [CrossRef] [Green Version]
- Hernaez, B.; Cabezas, M.; Munoz-Moreno, R.; Galindo, I.; A. Cuesta-Geijo, M.; Alonso, C. A179L, a New Viral Bcl2 Homolog Targeting Beclin 1 Autophagy Related Protein. Curr. Mol. Med. 2013, 13, 305–316. [Google Scholar] [CrossRef]
- Banjara, S.; Caria, S.; Dixon Linda, K.; Hinds Mark, G.; Kvansakul, M.; McFadden, G. Structural Insight into African Swine Fever Virus A179L-Mediated Inhibition of Apoptosis. J. Virol. 2017, 91, e02228-16. [Google Scholar] [CrossRef] [Green Version]
- Youle, R.J.; Strasser, A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef]
- Banjara, S.; Shimmon, G.L.; Dixon, L.K.; Netherton, C.L.; Hinds, M.G.; Kvansakul, M. Crystal Structure of African Swine Fever Virus A179L with the Autophagy Regulator Beclin. Viruses 2019, 11, 789. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Xie, M.; Wu, W.; Chen, Z. Structures and Functional Diversities of ASFV Proteins. Viruses 2021, 13, 2124. https://doi.org/10.3390/v13112124
Wang G, Xie M, Wu W, Chen Z. Structures and Functional Diversities of ASFV Proteins. Viruses. 2021; 13(11):2124. https://doi.org/10.3390/v13112124
Chicago/Turabian StyleWang, Guoguo, Mengjia Xie, Wei Wu, and Zhongzhou Chen. 2021. "Structures and Functional Diversities of ASFV Proteins" Viruses 13, no. 11: 2124. https://doi.org/10.3390/v13112124
APA StyleWang, G., Xie, M., Wu, W., & Chen, Z. (2021). Structures and Functional Diversities of ASFV Proteins. Viruses, 13(11), 2124. https://doi.org/10.3390/v13112124






