Application of the Fuzzy Oil Drop Model Describes Amyloid as a Ribbonlike Micelle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data
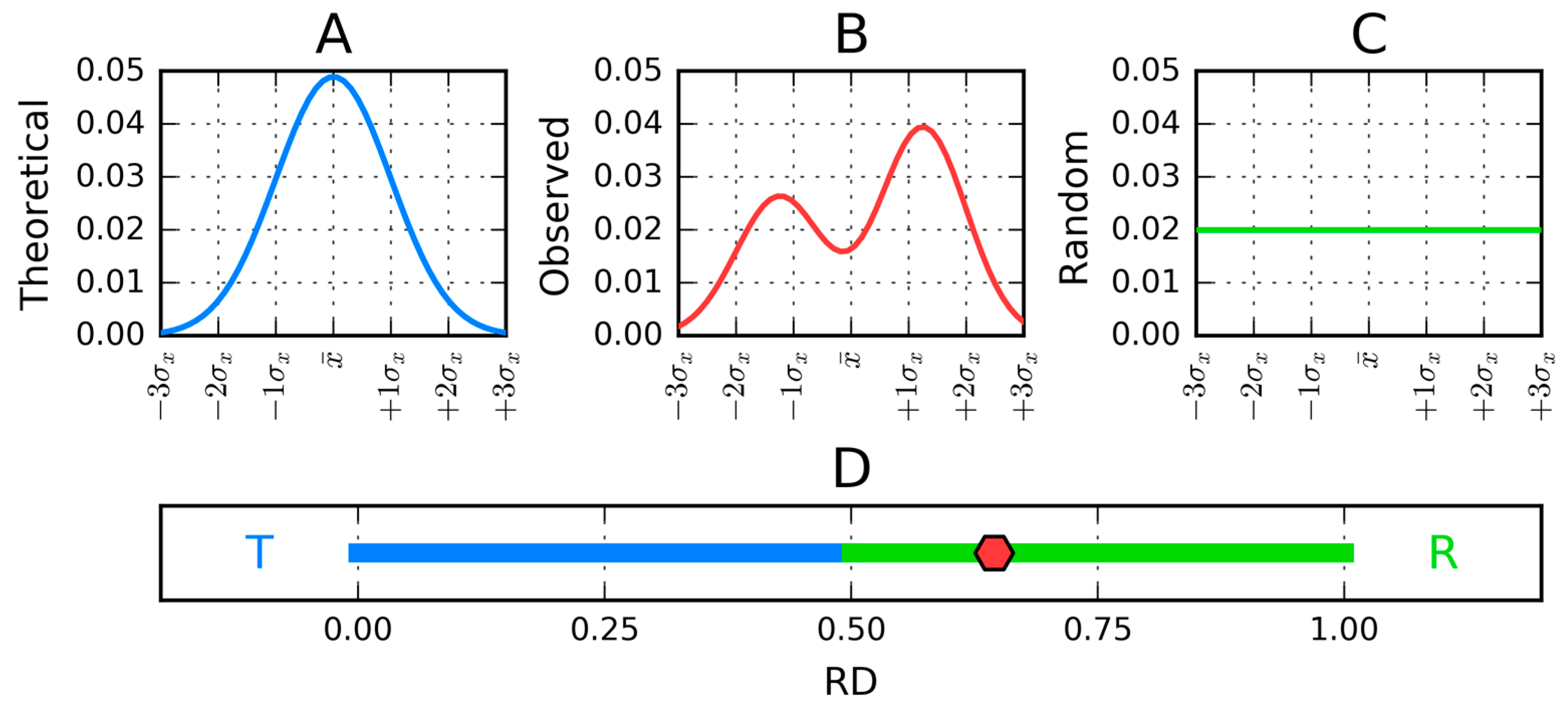
2.2. Fuzzy Oil Drop Model
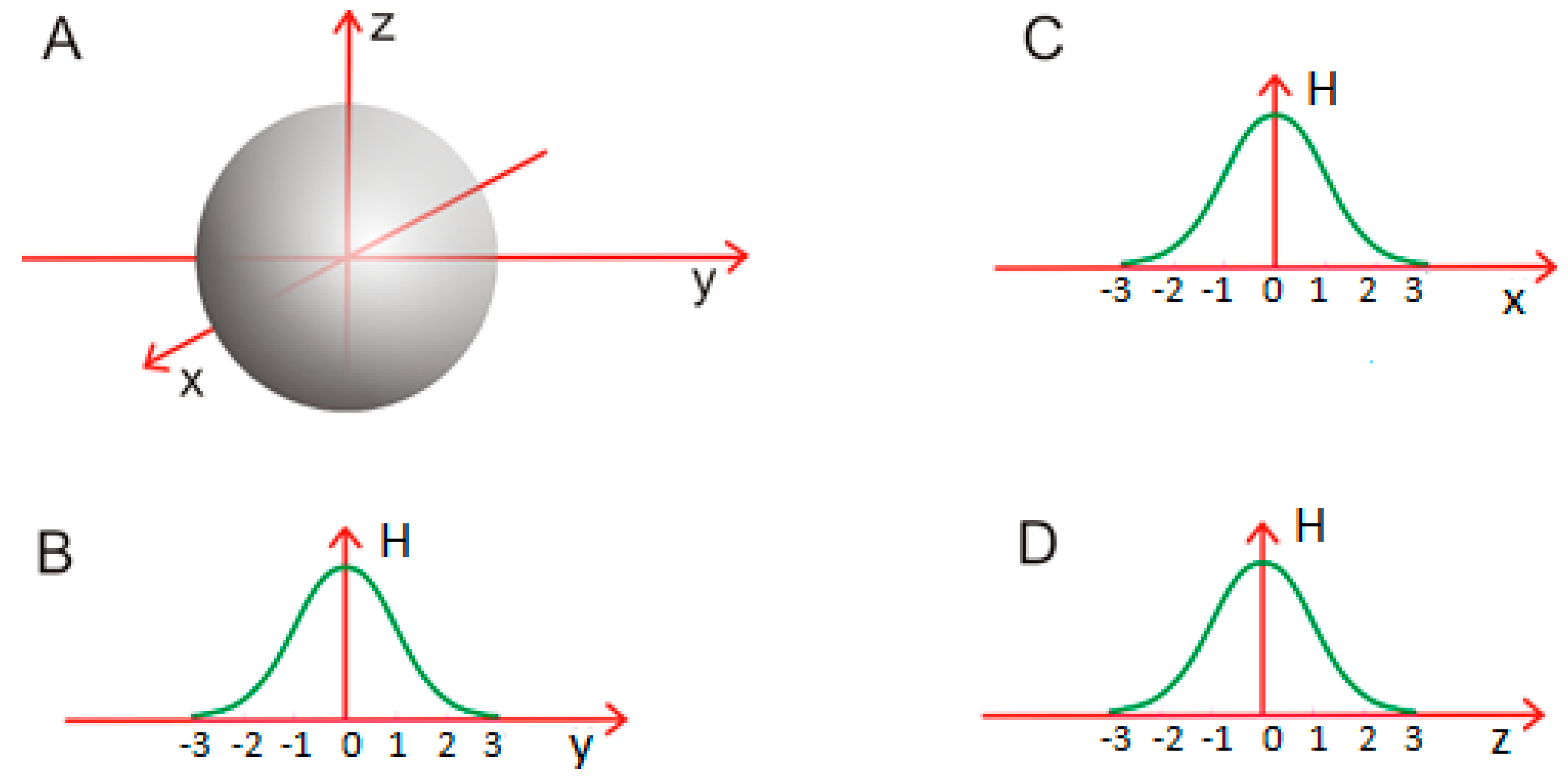
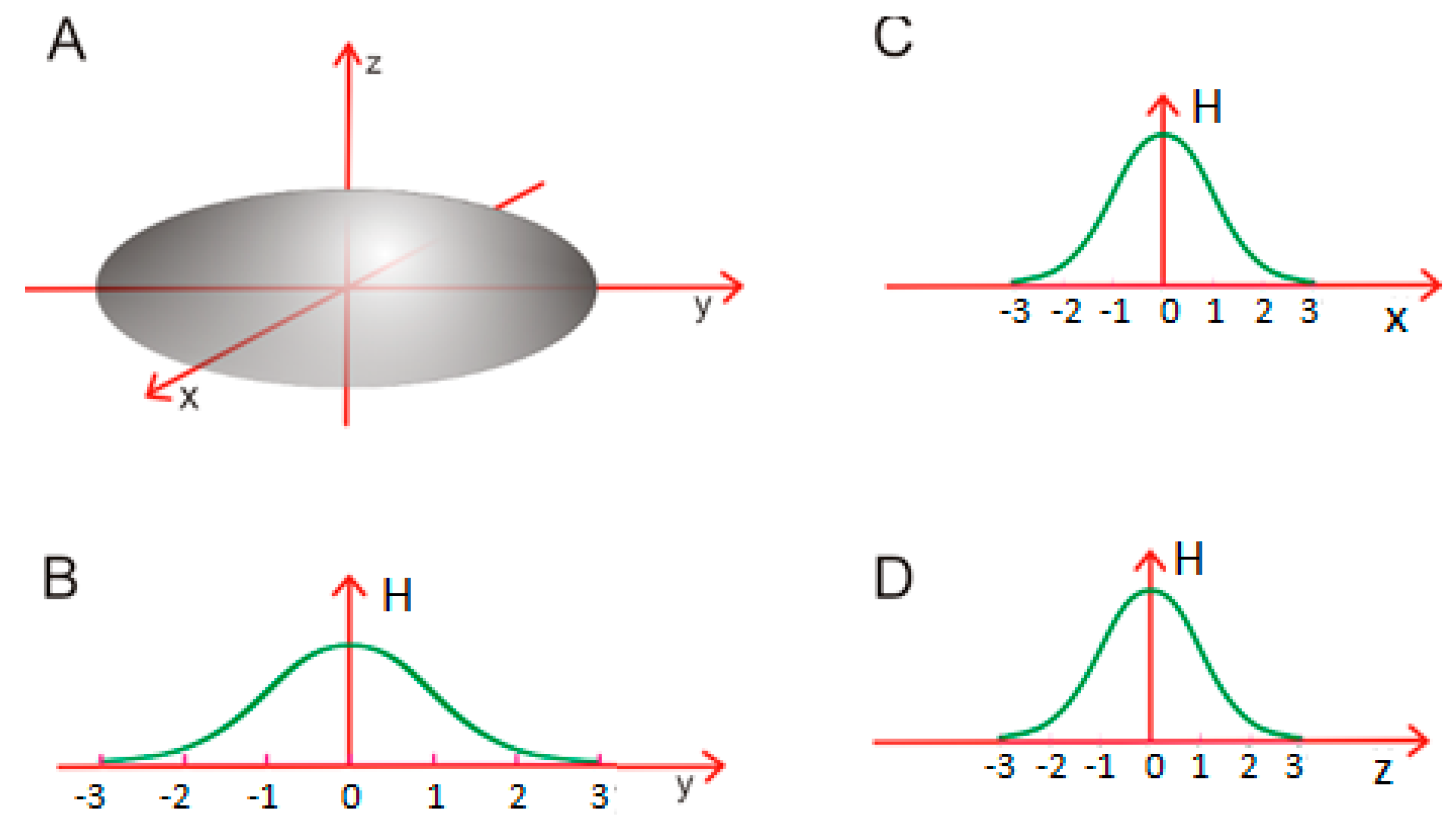
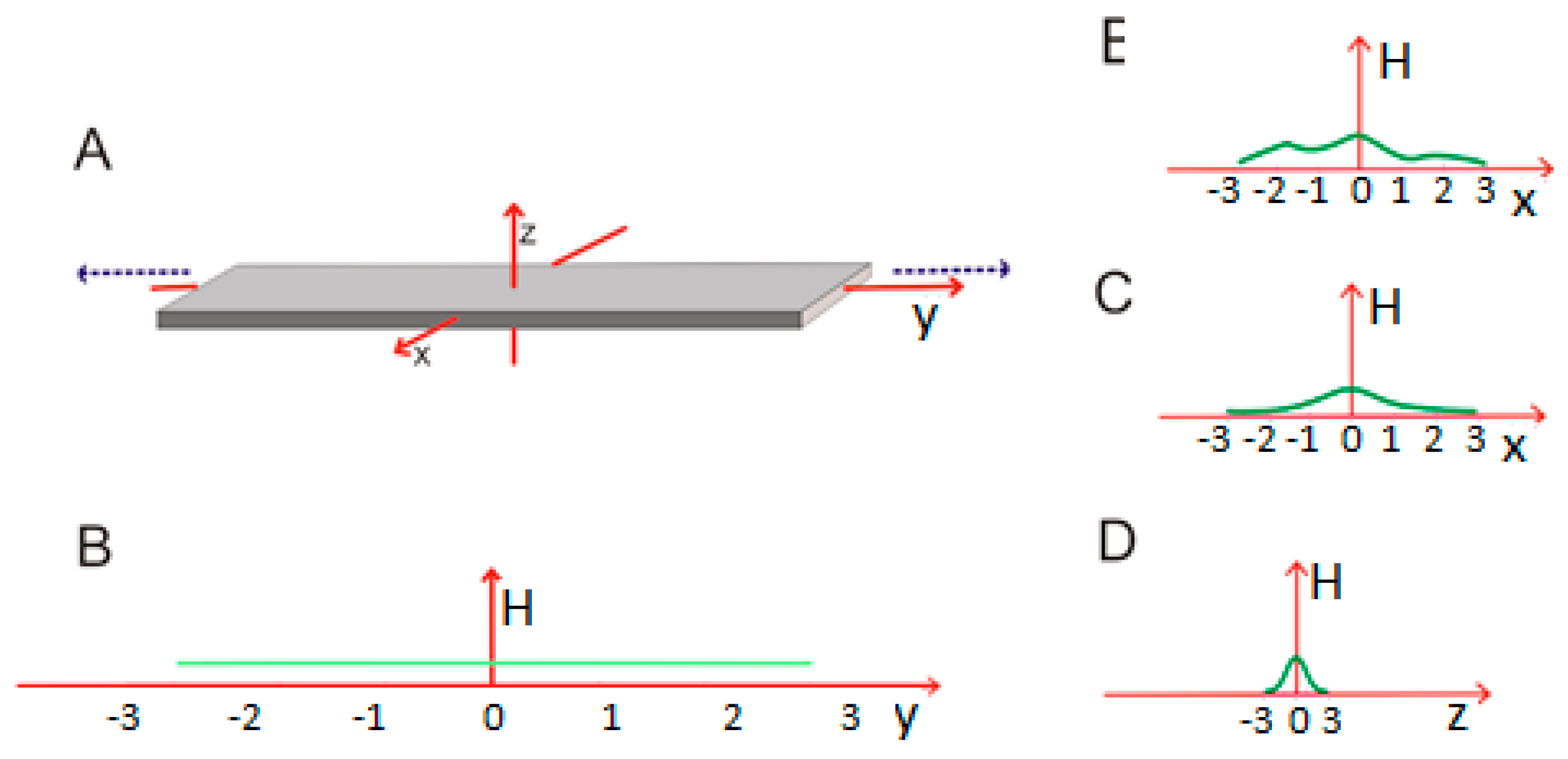
2.3. Modeling the Micellar Structure with a 3D Gaussian Function
3. Results
3.1. Distribution of Hydrophobicity in Short Segments of Amyloid-Forming Proteins
3.2. Amyloid Structure in the Context of the Fuzzy Oil Drop Model
3.3. Cylindrical Micelle in Proteins
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Prusiner, S.B. Novel proteinaceous infectious particles cause scrapie. Science 1982, 216, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B. Detecting madcow disease. Sci. Am. 2004, 291, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Stöhr, J.; Watts, J.C.; Mensinger, Z.L.; Oehler, A.; Grillo, S.K.; DeArmond, S.J.; Prusiner, S.B.; Giles, K. Purified and synthetic Alzheimer’s amyloid β (Aβ) prions. Proc. Natl. Acad. Sci. USA 2012, 109, 11025–11030. [Google Scholar] [CrossRef] [PubMed]
- Taubes, G. Misfolding the way to disease. Science 1996, 271, 1493–1495. [Google Scholar] [CrossRef] [PubMed]
- Purkey, H.E.; Dorrell, M.I.; Kelly, J.W. Evaluating the binding selectivity of transthyretin amyloid fibril inhibitors in blood plasma. Proc. Natl. Acad. Sci. USA 2001, 98, 5566–5571. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, R.; Ramakers, M.; De Smet, F.; Claes, F.; Khodaparast, L.; Khodaparast, L.; Couceiro, J.R.; Langenberg, T.; Siemons, M.; Nyström, S.; et al. De novo design of a biologically active amyloid. Science 2016, 354, 6313. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, L.; Bryliński, M.; Roterman, I. Gauss-function-based model of hydrophobicity density in proteins. In Silico Biol. 2006, 6, 15–22. [Google Scholar] [PubMed]
- Banach, M.; Konieczny, L.; Roterman, I. Ligand-binding site recognition. In Protein Folding in Silico—Protein Folding Versus Protein Structure Prediction; Roterman, I., Ed.; Woodhead Publishing: Cambridge, UK, 2012; pp. 79–94. [Google Scholar]
- Banach, M.; Konieczny, L.; Roterman, I. Use of the “fuzzy oil drop” model to identify the complexation area in protein homodimers. In Protein Folding in Silico—Protein Folding Versus Protein Structure Prediction; Roterman, I., Ed.; Woodhead Publishing: Cambridge, UK, 2012; pp. 95–122. [Google Scholar]
- Dygut, J.; Kalinowska, B.; Banach, M.; Piwowar, M.; Konieczny, L.; Roterman, I. Structural Interface Forms and Their Involvement in Stabilization of Multidomain Proteins or Protein Complexes. Int. J. Mol. Sci. 2016, 17, 1741. [Google Scholar] [CrossRef] [PubMed]
- Versace, R.E.; Lazaridis, T. Modeling Protein–Micelle Systems in Implicit Water. J. Phys. Chem. B 2015, 119, 8037–8047. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C.; Hristova, K.; Ladokhin, A.S.; Silvestro, L.; Axelsen, P.H.; White, S.H. Folding of β-sheet membrane proteins: A hydrophobic hexapeptide model. J. Mol. Biol. 1998, 277, 1091–1110. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolo, N.; Compton, E.L.; Warne, T.; Edwards, P.C.; Tate, C.G.; Schertler, G.F.; Booth, P.J. Complete Reversible Refolding of a G-Protein Coupled Receptor on a Solid Support. PLoS ONE 2016, 11, e0151582. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Xiang, L.; Liu, Y.; Deng, X.; Cao, X.; Li, L.; Yu, C.; Chen, Y.; Tang, G. Synthesis and in vitro evaluation of pH-sensitive PEG-I-dC16 block polymer micelles for anticancer drug delivery. J. Pharm. Pharmacol. 2016, 68, 751–761. [Google Scholar]
- Jaskula-Sztul, R.; Xu, W.; Chen, G.; Harrison, A.; Dammalapati, A.; Nair, R.; Cheng, Y.; Gong, S.; Chen, H. Thailandepsin A-loaded and octreotide-functionalized unimolecular micelles for targeted neuroendocrine cancer therapy. Biomaterials 2016, 91, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bar-Zeev, M.; Assaraf, Y.; Livney, Y.D. β-casein nanovehicles for oral delivery of chemotherapeutic Drug combinations overcoming P-glycoprotein-mediated multidrug resistance in human gastric cancer cells. Oncotarget 2016, 7, 23322–23334. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Miyamoto, Y.; Cabral, H.; Matsumoto, Y.; Nagasaka, K.; Nakagawa, S.; Yano, T.; Maeda, D.; Oda, K.; Kawana, K.; et al. Enhanced efficacy against cervical carcinomas through polymeric micelles physically incorporating the proteasome inhibitor MG132. Cancer Sci. 2016, 107, 773–7781. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Youn, Y.S.; Lee, K.S.; Hoang, N.H.; Sim, T.; Lee, E.S.; Oh, K.T. Development of a robust pH-sensitive polyelectrolyte ionomer complex for anticancer nanocarriers. Int. J. Nanomed. 2016, 11, 703–713. [Google Scholar]
- Chren, Y.; Huang, Y.; Qin, D.; Liu, W.; Song, C.; Lou, K.; Wang, W.; Gao, F. β-Cyclodextrin-Based Inclusion Complexation Bridged Biodegradable Self-Assembly Macromolecular Micelle for the Delivery of Paclitaxel. PLoS ONE 2016, 11, e0150877. [Google Scholar] [CrossRef]
- Karaca, M.; Dutta, R.; Ozsoy, Y.; Mahato, R.I. Micelle Mixtures for Coadministration of Gemcitabine and GDC-0449 to Treat Pancreatic Cancer. Mol. Pharm. 2016, 13, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Tiana, G.; Simona, F.; Broglia, R.A.; Colombo, G. Thermodynamics of β-amyloid fibril formation. J. Chem. Phys. 2004, 120, 8307–8317. [Google Scholar] [CrossRef] [PubMed]
- Serra-Batiste, M.; Ninot-Pedrosa, M.; Bayoumi, M.; Gairí, M.; Maglia, G.; Carulla, N. Aβ42 assembles into specific β-barrel pore-forming oligomers in membrane-mimicking environments. Proc. Natl. Acad. Sci. USA 2016, 113, 10866–10871. [Google Scholar] [CrossRef] [PubMed]
- So, M.; Ishii, A.; Hata, Y.; Yagi, H.; Naiki, H.; Goto, Y. Supersaturation-Limited and Unlimited Phase Spaces Compete to Produce Maximal Amyloid Fibrillation near the Critical Micelle Concentration of Sodium Dodecyl Sulfate. Langmuir 2015, 31, 9973–9982. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Kobayashi, K.; Maeda, T.; Sato, K.; Takashima, A. Three-dimensional structures of the amyloid β peptide (25–35) in membrane-mimicking environment. Biochemistry 1996, 35, 16094–16104. [Google Scholar] [CrossRef] [PubMed]
- Riek, R.; Eisenberg, D.S. The activities of amyloids from a structural perspective. Nature 2016, 339, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Ma, B.; McElheny, D.; Parthasarathy, S.; Long, F.; Hoshi, M.; Nussinov, R.; Ishii, Y. Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat. Struct. Mol. Biol. 2015, 22, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Matte, A.; Li, Y.; Kim, Y.S.; Linhardt, R.J.; Su, H.; Cygler, M. Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution. J. Mol. Biol. 1999, 294, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.; Sawaya, M.R.; Balbirnie, M.; Madsen, A.Ø.; Riekel, C.; Grothe, R.; Eisenberg, D. Structure of the cross-β spine of amyloid-like fibrils. Nature 2005, 435, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Colletier, J.P.; Laganowsky, A.; Landau, M.; Zhao, M.; Soriaga, A.B.; Goldschmidt, L.; Flot, D.; Cascio, D.; Sawaya, M.R.; Eisenberg, D. Molecular basis for amyloid-β polymorphism. Proc. Natl. Acad. Sci. USA 2011, 108, 16938–16943. [Google Scholar] [CrossRef] [PubMed]
- Wiltzius, J.J.; Landau, M.; Nelson, R.; Sawaya, M.R.; Apostol, M.I.; Goldschmidt, L.; Soriaga, A.B.; Cascio, D.; Rajashankar, K.; Eisenberg, D. Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 2009, 16, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Sawaya, M.R.; Eisenberg, D. β2-microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat. Struct. Mol. Biol. 2011, 18, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Apostol, M.I.; Wiltzius, J.J.; Sawaya, M.R.; Cascio, D.; Eisenberg, D. Atomic structures suggest determinants of transmission barriers in mammalian prion disease. Biochemistry 2011, 50, 2456–2463. [Google Scholar] [CrossRef] [PubMed]
- Apostol, M.; Sawaya, M.R.; Cascio, D.; Eisenberg, D. Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease. J. Biol. Chem. 2010, 285, 29671–29675. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Charles, I.G.; Fairweather, N.F.; Isaacs, N.W. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature 1996, 381, 90–92. [Google Scholar] [CrossRef] [PubMed]
- Kalinowska, B.; Banach, M.; Konieczny, L.; Roterman, I. Application of Divergence Entropy to Characterize the Structure of the Hydrophobic Core in DNA Interacting Proteins. Entropy 2015, 17, 1477–1507. [Google Scholar] [CrossRef]
- Roterman, I.; Banach, M.; Kalinowska, B.; Konieczny, L. Influence of the Aqueous Environment on Protein Structure—A Plausible Hypothesis Concerning the Mechanism of Amyloidogenesis. Entropy 2016, 18, 351. [Google Scholar] [CrossRef]
- Kullback, S.; Leibler, R.A. On information and sufficiency. Ann. Math. Stat. 1951, 22, 79–86. [Google Scholar] [CrossRef]
- Król, M.; Borowski, T.; Roterman, I.; Piekarska, B.; Stopa, B.; Rybarska, J.; Konieczny, L. Force-field parametrization and molecular dynamics simulations of Congo red. J. Comput. Aided Mol. Des. 2004, 18, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Banach, M.; Prymula, K.; Jurkowski, W.; Konieczny, L.; Roterman, I. Fuzzy oil drop model to interpret the structure of antifreeze proteins and their mutants. J. Mol. Model. 2012, 18, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Roterman, I.; Konieczny, L.; Jurkowski, W.; Prymula, K.; Banach, M. Two-intermediate model to characterize the structure of fast-folding proteins. J. Theor. Biol. 2011, 283, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Wada, K.; Sumi, N.; Nagai, R.; Iwasaki, K.; Sato, T.; Suzuki, K.; Hasegawa, Y.; Kitaoka, S.; Minami, Y.; Outten, F.W.; et al. Molecular dynamism of fe-S cluster biosynthesis implicated by the structure of the sufc2-Sufd2 complex. J. Mol. Biol. 2009, 387, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X. Protein structure determination by solid-state NMR. Top. Curr. Chem. 2012, 326, 187–213. [Google Scholar] [PubMed]
- Wang, S.; Munro, R.A.; Shi, L.; Kawamura, I.; Okitsu, T.; Wada, A.; Kim, S.-Y.; Hwan, K.; Jung, K.-H.; Brown, L.-S.; et al. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat. Methods 2013, 10, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Roterman, I.; Banach, M.; Konieczny, L. Stop signals preventing the linear propagation of hydrophobicity in proteins. IJMS 2017. submitted. [Google Scholar]
- Banach, M.; Konieczny, L.; Roterman, I. The fuzzy oil drop model, based on hydrophobicity density distribution, generalizes the influence of water environment on protein structure and function. J. Theor. Biol. 2014, 359, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Levinthal, C. Are there pathways for protein folding. J. Chem. Phys. 1968, 65, 44–45. [Google Scholar]
- Tanford, C. Protein denaturation. Adv. Protein Chem. 1968, 23, 121–282. [Google Scholar] [PubMed]
- Agashe, V.R.; Shastry, M.C.; Udgaonkar, J.B. Initial hydrophobic collapse in the folding of barstar. Nature 1995, 377, 754–757. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Kollman, P.A. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science 1998, 282, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Burke, M.G.; Woscholski, R.; Yaliraki, S.N. Differential hydrophobicity drives self-assembly in Huntington’s disease. Proc. Natl. Acad. Sci. USA 2003, 100, 13928–13933. [Google Scholar] [CrossRef] [PubMed]
- Jurkowski, W.; Brylinski, M.; Konieczny, L.; Wiíniowski, Z.; Roterman, I. Conformational subspace in simulation of early-stage protein folding. Proteins 2004, 55, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Serpell, L.C. Alzheimer’s amyloid fibrils: Structure and assembly. Biochim. Biophys. Acta 2000, 1502, 16–30. [Google Scholar] [CrossRef]
- Biedermann, F.; Nau, W.M.; Schneider, H.-J. The hydrophobic effect revisited—Studies with supramolecular complexes imply high-energy water as a noncovalent driving force. Angew. Chem. 2014, 53, 11158–11171. [Google Scholar] [CrossRef] [PubMed]
- Ben-Naim, A. Solvent-induced interactions: Hydrophobic and Hydrophilic Phenomena. J. Chem. Phys. 1989, 90, 7412–7525. [Google Scholar] [CrossRef]
- Grimaldi, M.; Scrima, M.; Esposito, C.; Vitiello, G.; Ramunno, A.; Limongelli, V.; D’Errico, G.; Novellino, E.; D’Ursi, A.M. Membrane charge dependent states of the β-amyloid fragment Aβ (16–35) with differently charged micelle aggregates. Biochim. Biophys. Acta 2010, 1798, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, E.; Gafni, A. Micelle formation by a fragment of human islet amyloid polypeptide. Biophys. J. 2003, 84, 3480–3487. [Google Scholar] [CrossRef]
- Bisaglia, M.; Trolio, A.; Bellanda, M.; Bergantino, E.; Bubacco, L.; Mammi, S. Structure and topology of the non-amyloid-β component fragment of human alpha-synuclein bound to micelles: Implications for the aggregation process. Protein Sci. 2006, 15, 1408–1416. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Hu, D.; Zhu, M.; Fink, A.L. Structural chracterisation of the partially folder intermediates of an immunoglobulin Ligot chain leading to amyloid fibrillation and amorphous aggregation. Biochemistry 2007, 46, 3521–3531. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Folding proteins in fatal ways. Nature 2003, 426, 900–904. [Google Scholar] [CrossRef] [PubMed]

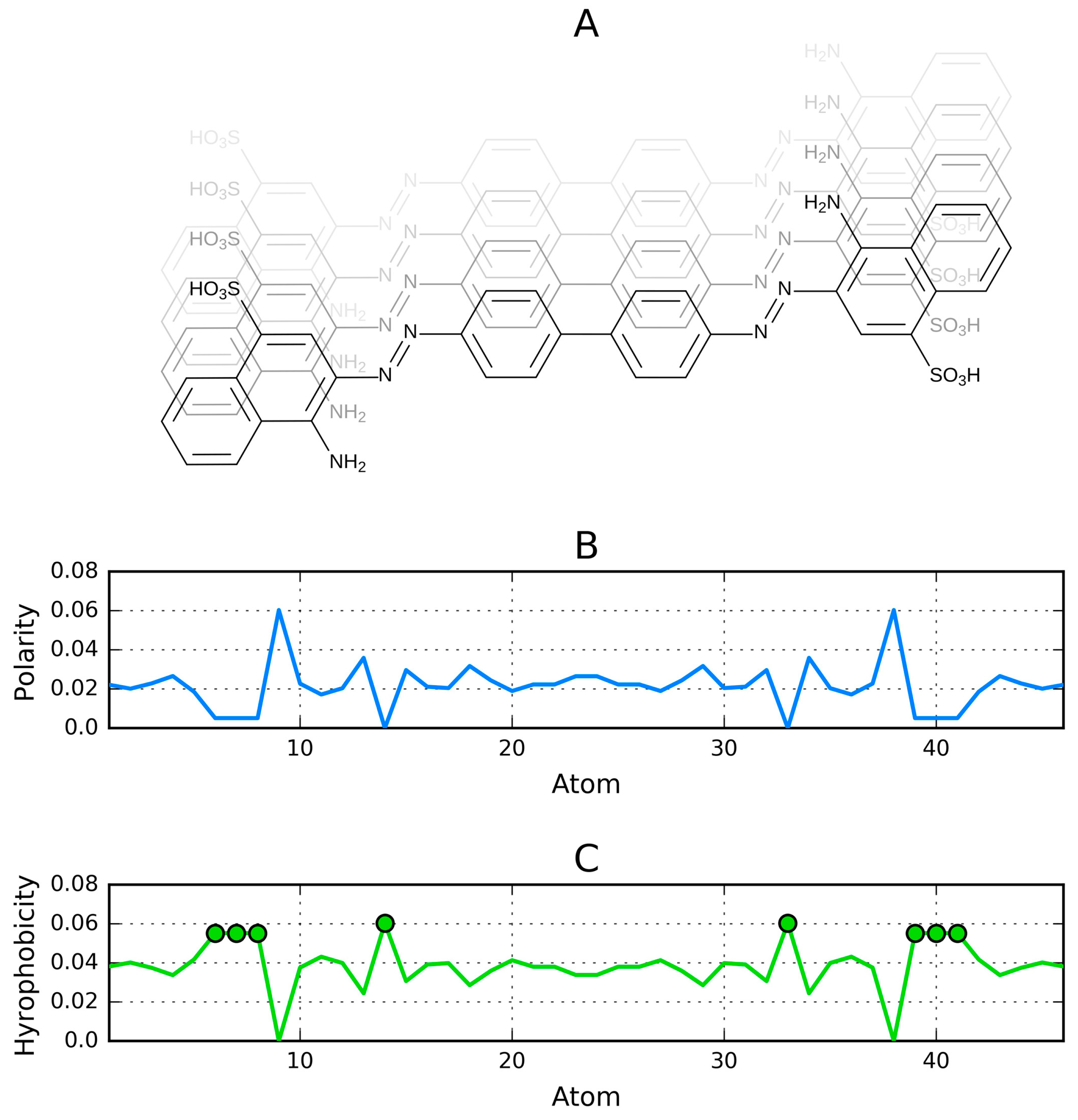
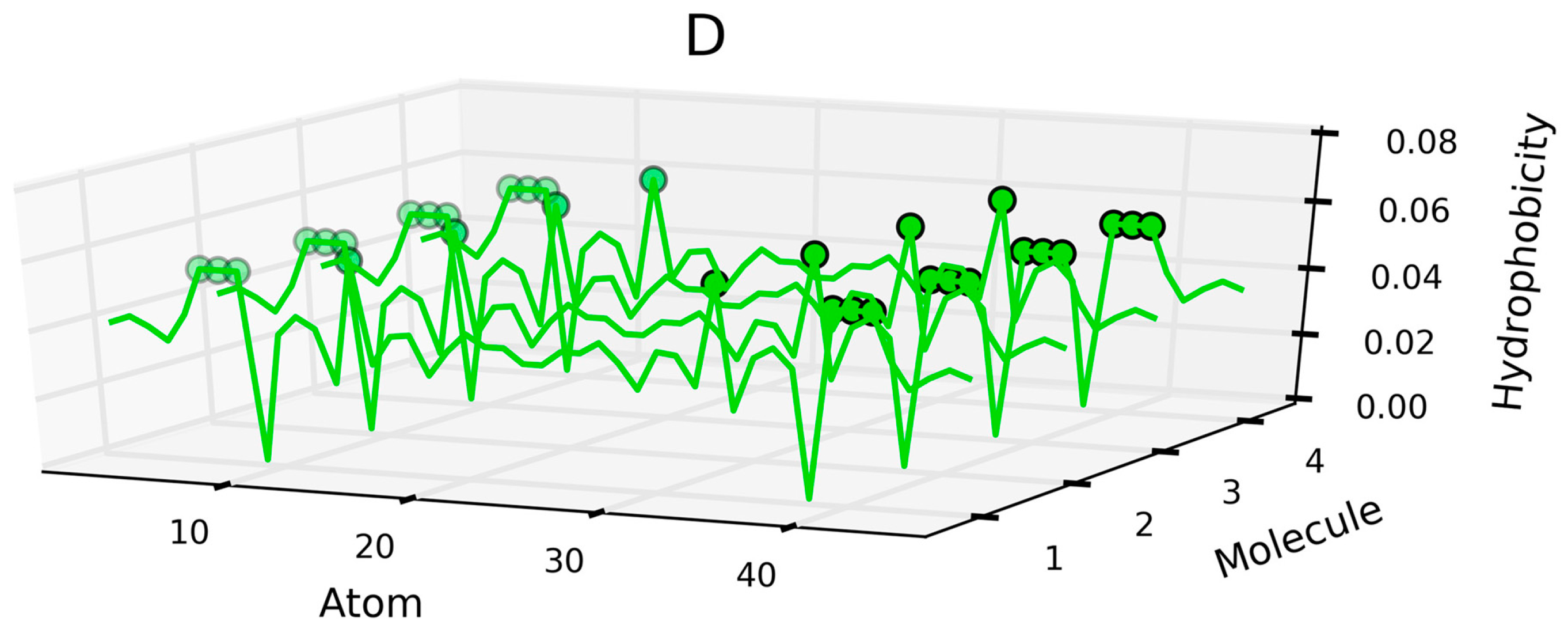
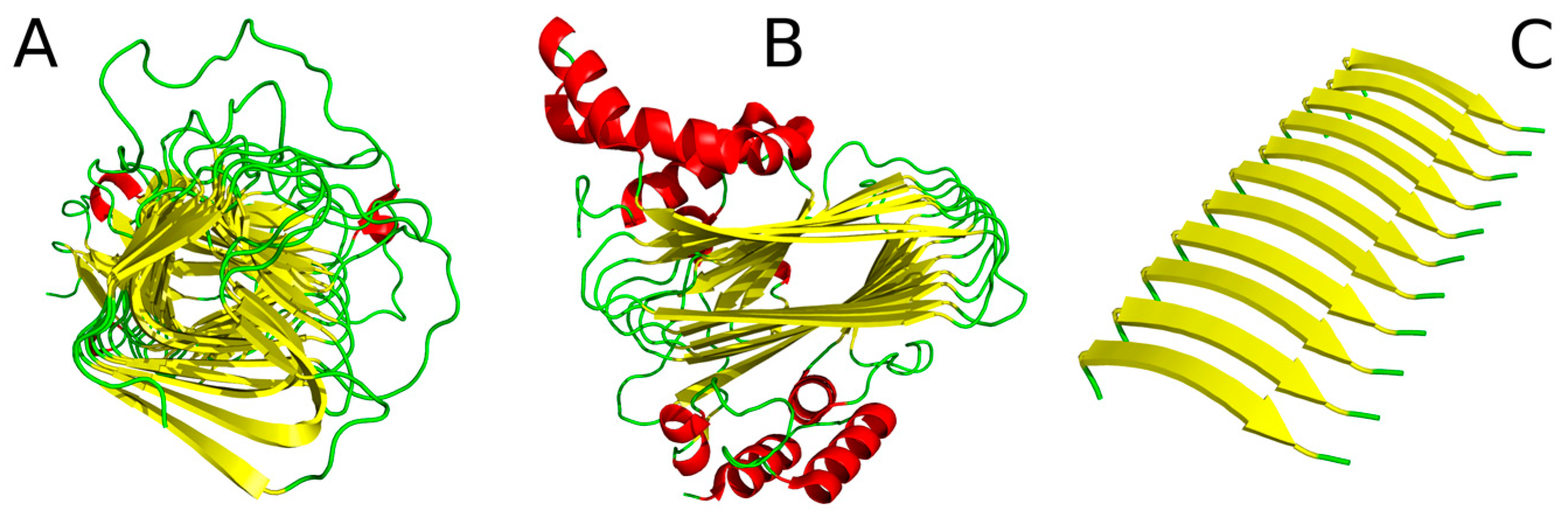

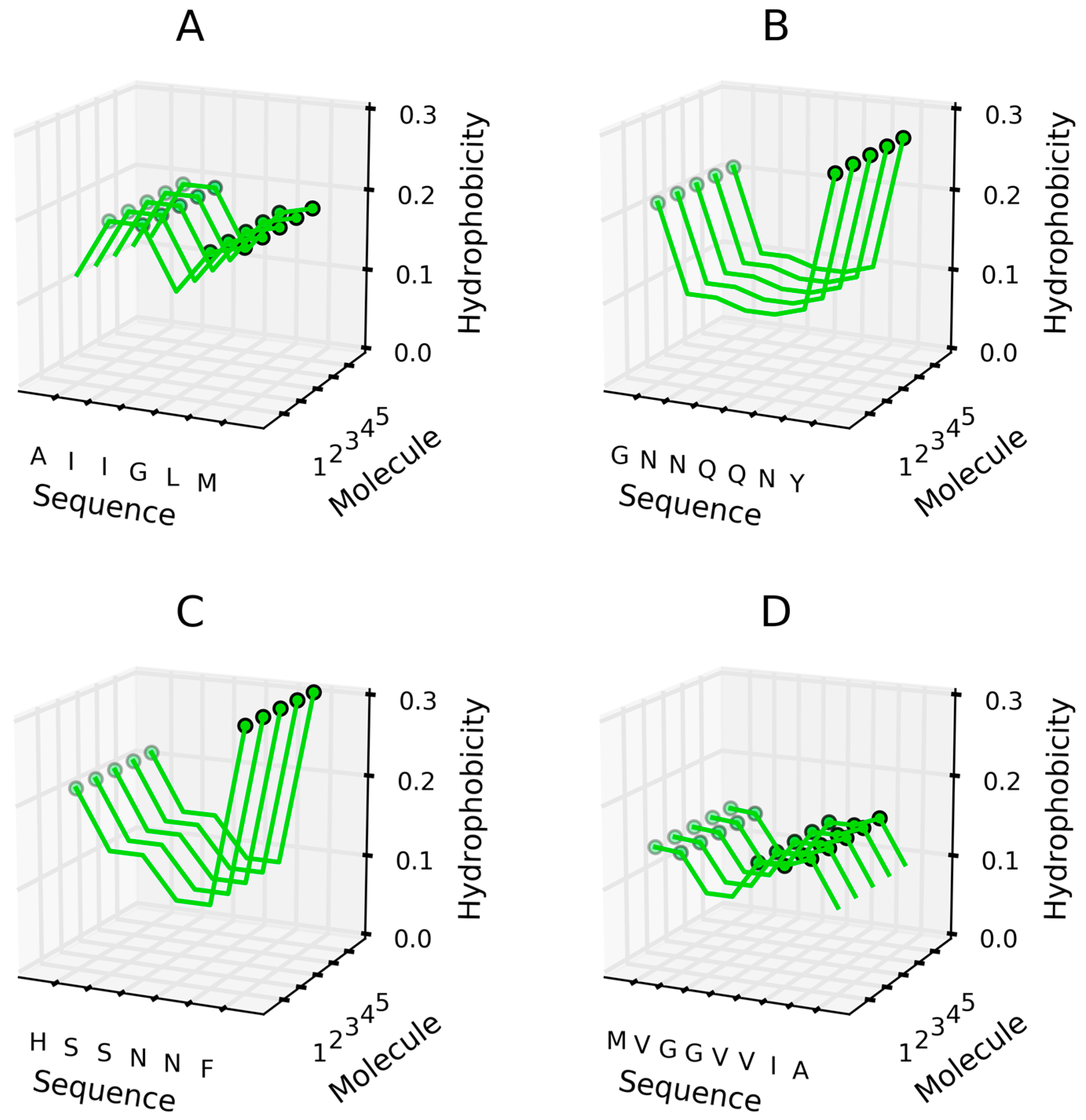

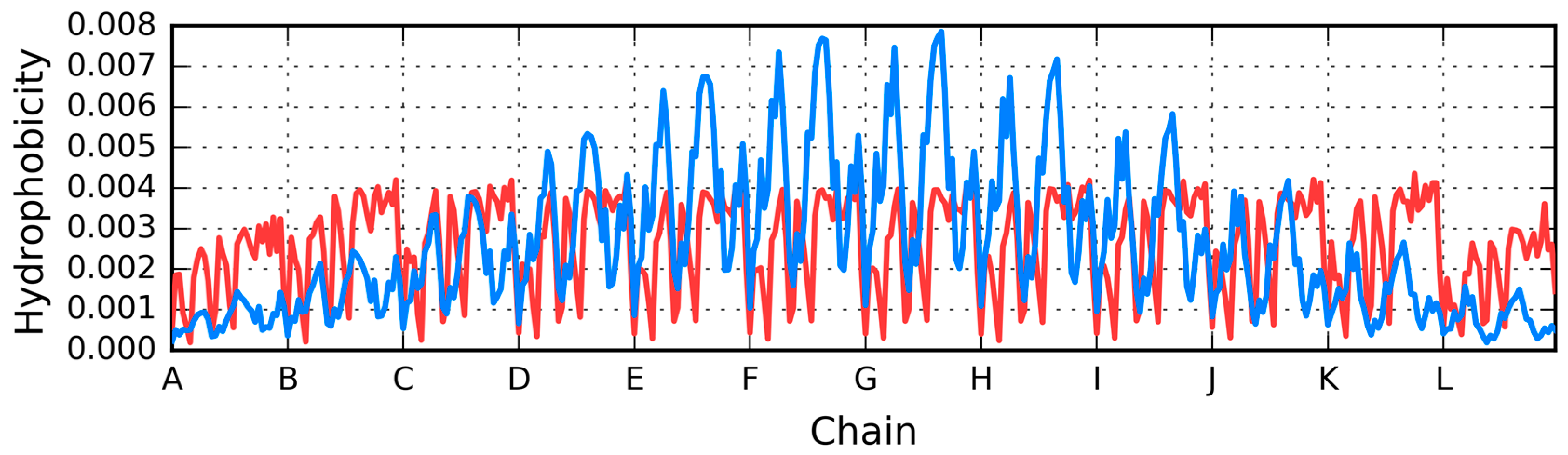
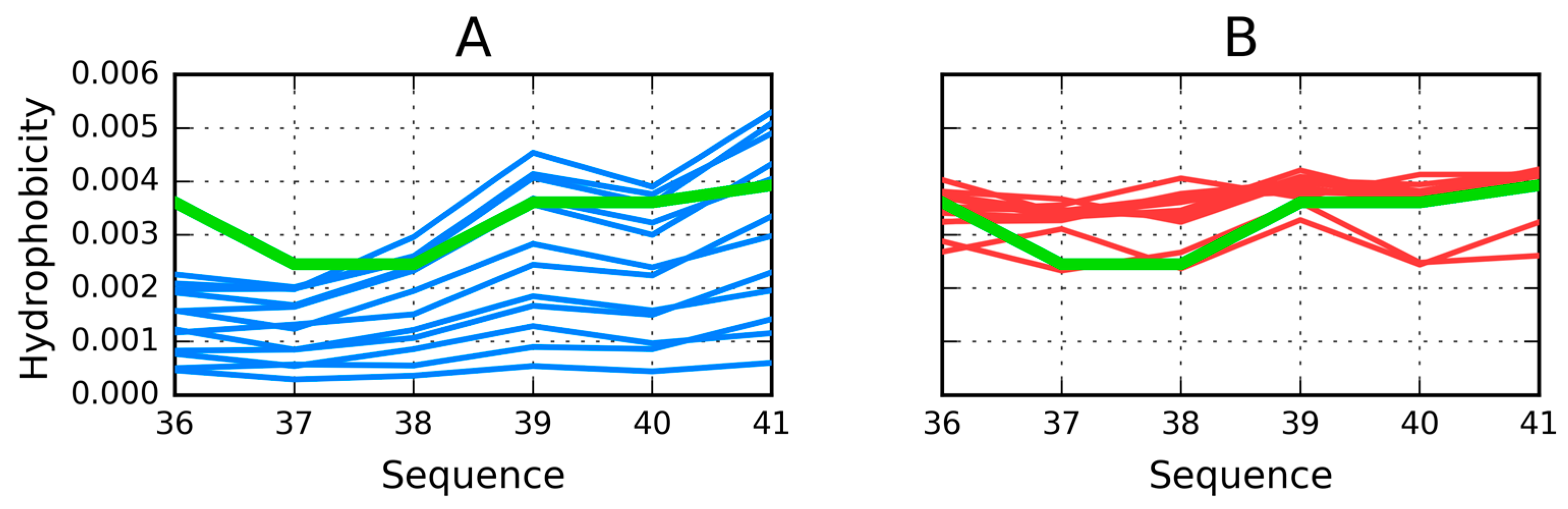
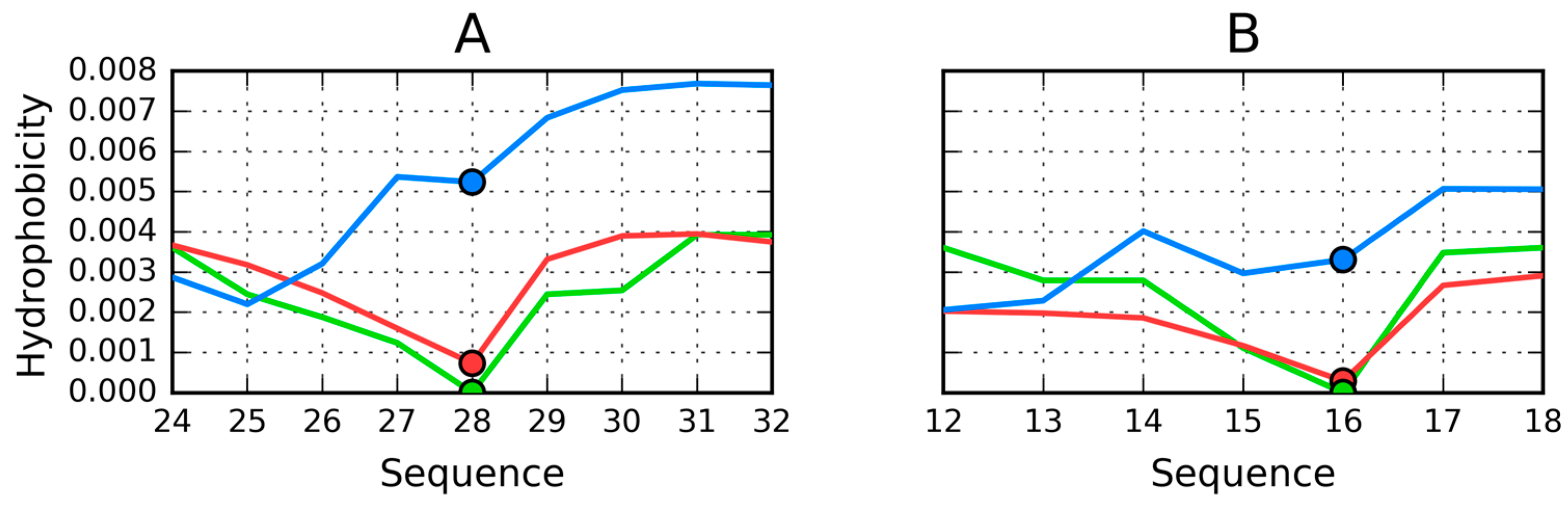
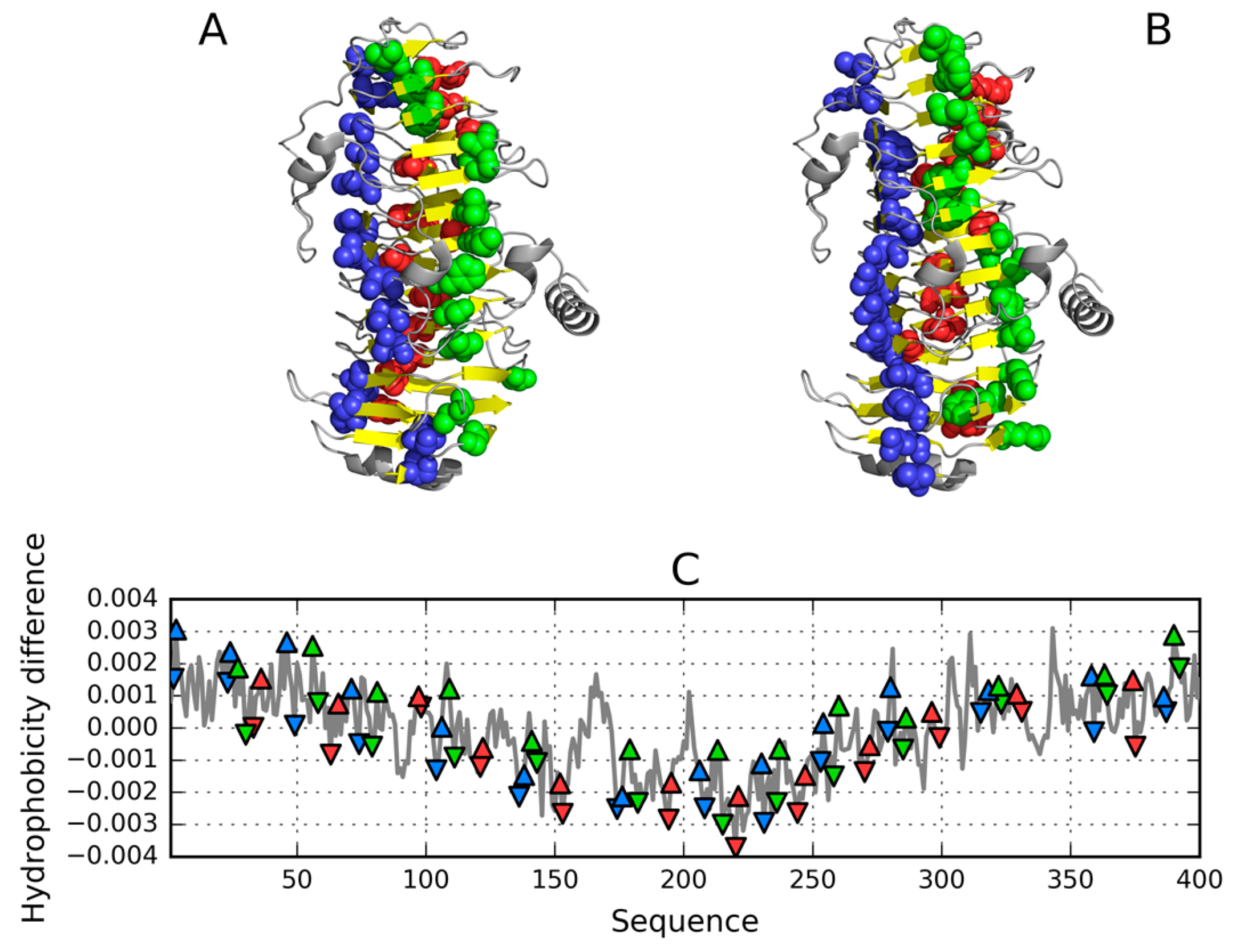
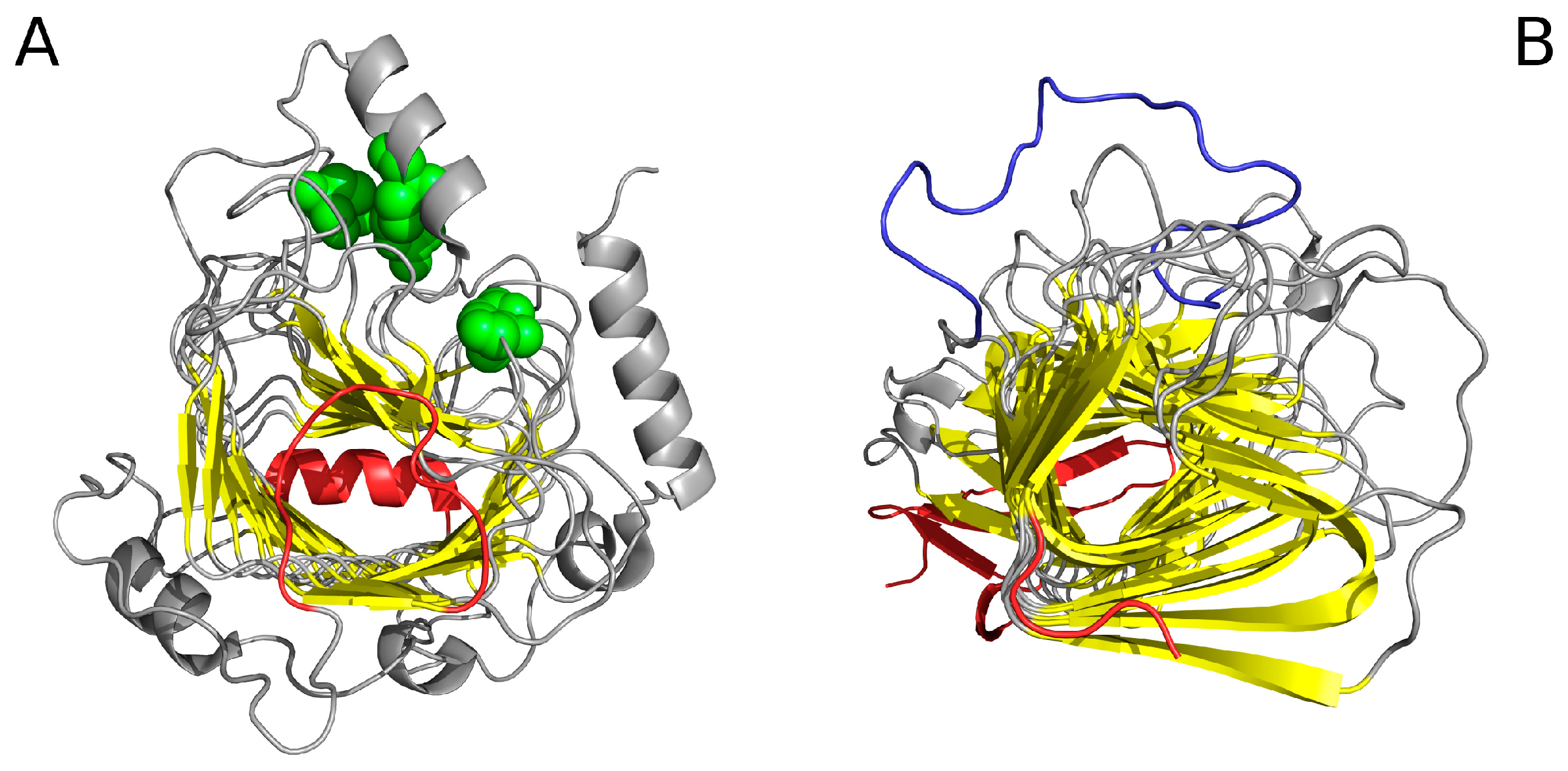
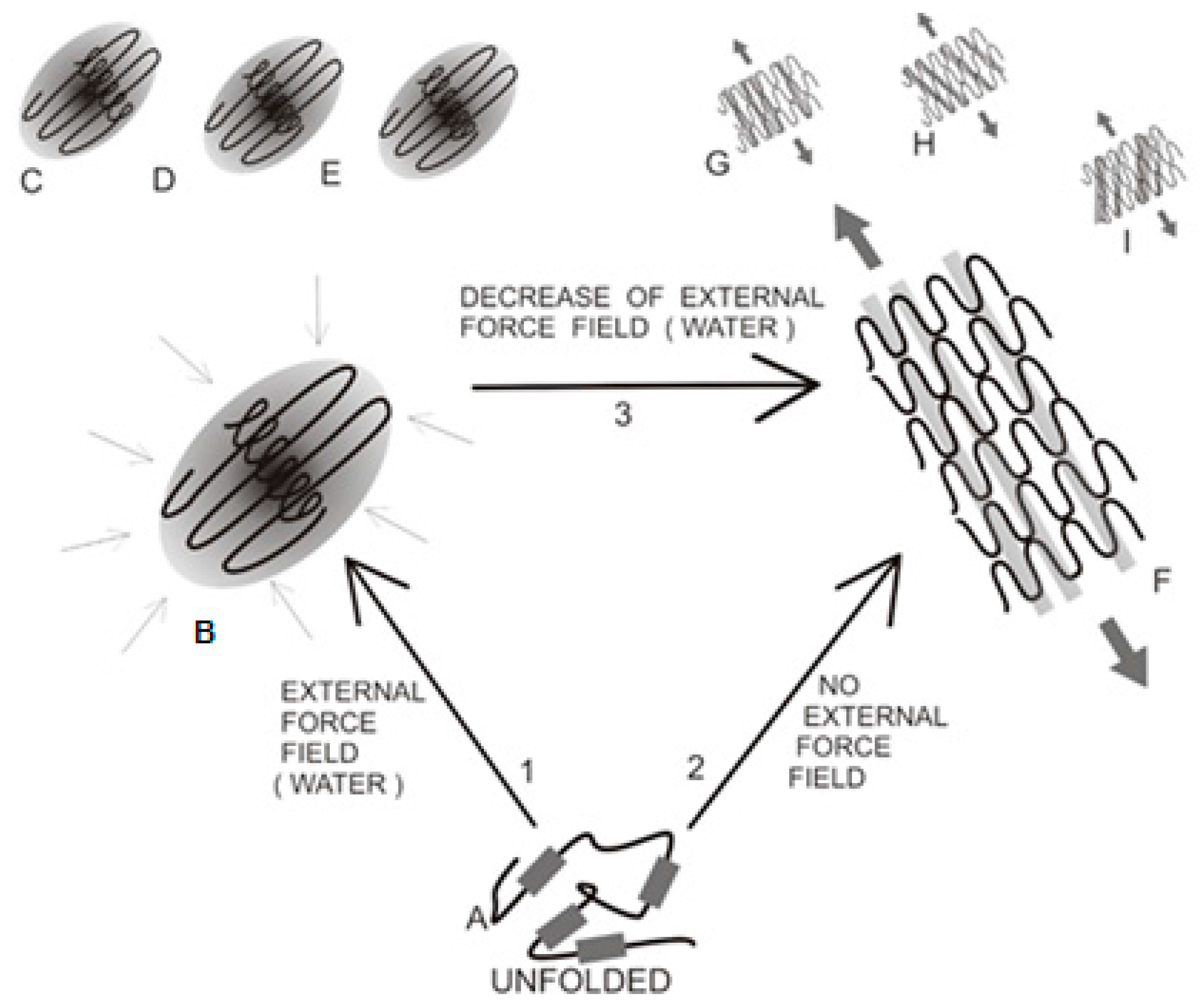
| Peptide Protein | Characteristics | Sequence | Characteristics | Reference |
|---|---|---|---|---|
| Ribbonlike Micelle | ||||
| 1YJP | prion | GNNQQNY | parallel | [27] |
| 2Y3J | amyloid beta | AIIGLM | parallel | [28] |
| 3FPO | Islet Amyloid polypeptide | HSSNNF | parallel | [29] |
| 3LOZ | macroglobulin | LSFSKD | antiparallel | [30] |
| 3NVE | prion | MMHFGN | antiparallel | [31] |
| 2Y3K | amyloid beta | MVGGVVIA | antiparallel | [32] |
| 3NHC | prion | GYMLGS | antiparallel | [29] |
| Cylindrical Micelle Composed of Ribbonlike Micelles | ||||
| 2MXU | human amyloid β (Aβ(1-42)) | 42 aa | parallel | [26] |
| Cylindrical Micelle | ||||
| 1DBG | Solenoid | parallel | [33] | |
| 1DAB | Solenoid | parallel | [27] | |
| RD | Correlation Coefficient | ||||
|---|---|---|---|---|---|
| 2MXU | RD(R) | RD(H) | ρ(H–T) | ρ(T–O) | ρ(H–O) |
| Complete | 0.680 | 0.756 | 0.246 | 0.364 | 0.821 |
| Chain | |||||
| A | 0.467 | 0.556 | 0.385 | 0.502 | 0.813 |
| B | 0.500 | 0.600 | 0.424 | 0.466 | 0.864 |
| C | 0.499 | 0.597 | 0.410 | 0.476 | 0.876 |
| D | 0.496 | 0.580 | 0.409 | 0.487 | 0.857 |
| E | 0.501 | 0.609 | 0.410 | 0.486 | 0.858 |
| F | 0.513 | 0.620 | 0.404 | 0.471 | 0.849 |
| G | 0.530 | 0.640 | 0.397 | 0.429 | 0.854 |
| H | 0.544 | 0.676 | 0.387 | 0.394 | 0.852 |
| I | 0.567 | 0.672 | 0.380 | 0.349 | 0.842 |
| J | 0.595 | 0.711 | 0.365 | 0.294 | 0.865 |
| K | 0.613 | 0.700 | 0.346 | 0.236 | 0.837 |
| L | 0.646 | 0.653 | 0.287 | 0.209 | 0.787 |
| β-sheet fragment | |||||
| 12–18 | 0.505 | 0.594 | 0.284 | 0.421 | 0.956 |
| 24–32 | 0.660 | 0.551 | 0.217 | 0.227 | 0.922 |
| 36–41 | 0.888 | 0.733 | 0.709 | 0.776 | 0.593 |
| β-sheet | |||||
| 12–18 | 0.655 | 0.726 | 0.136 | 0.329 | 0.920 |
| 24–32 | 0.770 | 0.684 | 0.169 | 0.265 | 0.896 |
| 36–41 | 0.947 | 0.902 | 0.997 | 0.998 | 0.999 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roterman, I.; Banach, M.; Konieczny, L. Application of the Fuzzy Oil Drop Model Describes Amyloid as a Ribbonlike Micelle. Entropy 2017, 19, 167. https://doi.org/10.3390/e19040167
Roterman I, Banach M, Konieczny L. Application of the Fuzzy Oil Drop Model Describes Amyloid as a Ribbonlike Micelle. Entropy. 2017; 19(4):167. https://doi.org/10.3390/e19040167
Chicago/Turabian StyleRoterman, Irena, Mateusz Banach, and Leszek Konieczny. 2017. "Application of the Fuzzy Oil Drop Model Describes Amyloid as a Ribbonlike Micelle" Entropy 19, no. 4: 167. https://doi.org/10.3390/e19040167






