Chemical Property Calculation through JavaScript and Applications in QSAR
Abstract
:Introduction
Calculation
| Molecular Weight Calculating Spreadsheet |
| http://www.unibas.ch/mdpi/ecsoc/e0002/mwcalcf.htm |
| logP Calculating using JavaScript |
| http://www.unibas.ch/mdpi/ecsoc/e0002/logpcalc.htm |
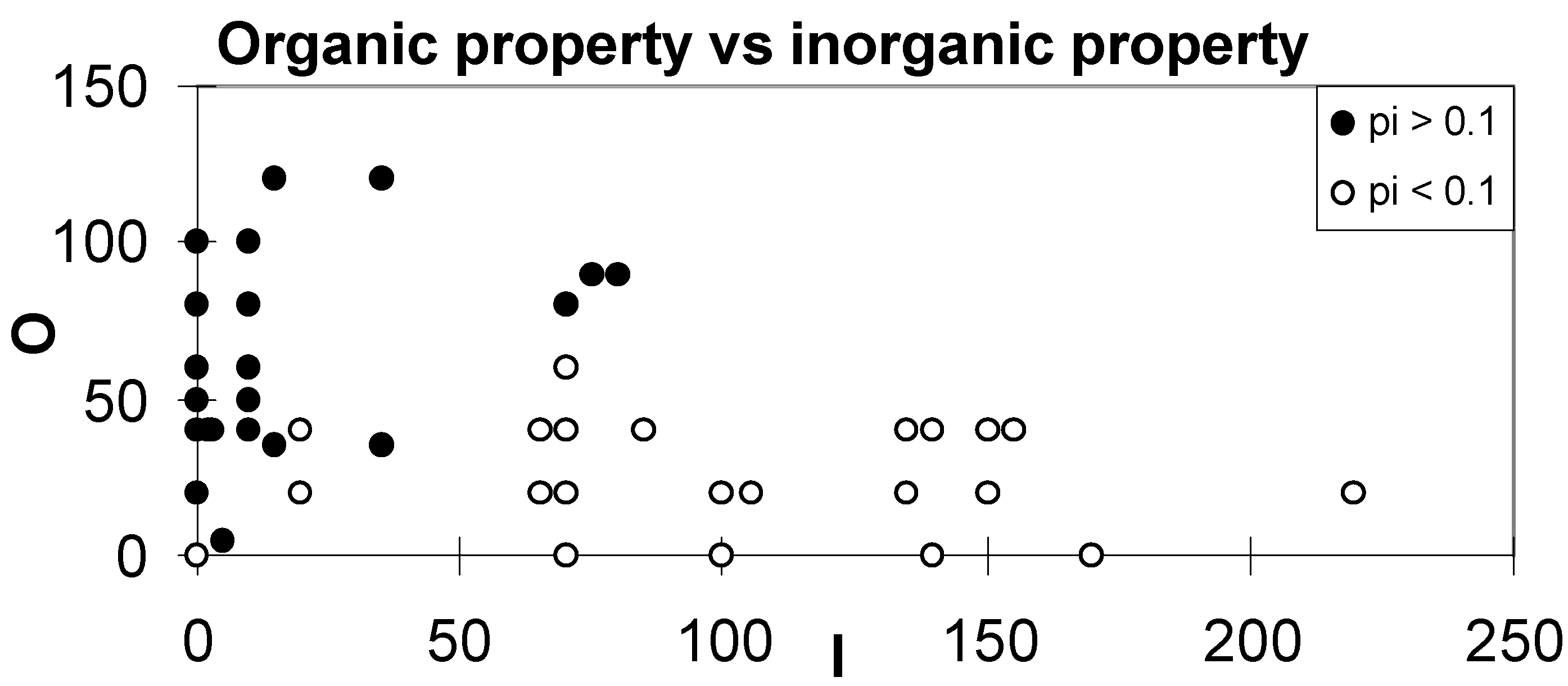
| Inorganic property and organic property values of groups. Calculation of Inorganic property value of molecules or groups using JavaScript. Calculation of Organic property value of molecules or groups using JavaScript. |
| Http://www.unibas.ch/mdpi/ecsoc/e0002/calinorg.htm |
| Calculation of Group Electronegativity using JavaScript. |
| Http://www.unibas.ch/mdpi/ecsoc/e0002/calelecg.htm |
Results
- logP = 5.0863 - 0.01618 I
- n = 20, r = 0.7420, s = 1.0423, F = 22.04
- logP = -1.5130 + 0.01710 O
- n = 20, r = 0.5256, s = 1.3226, F = 6.87
- logP = 3.3275 - 0.01685 I + 0.01847 O
- n = 20, r = 0.9337, s = 0.5729, F = 57.78
Applications
| H1 = I/O - 3, if H1 < 0, the amino acid is hydrophobic, otherwise hydrophilic; H2 = I - O - 160, if H2 < 0, the amino acid is hydrophobic, otherwise hydrophilic. |
| http://www.unibas.ch/mdpi/ecsoc/e0002/aa_io.htm |
+ 0.16273 MR1 + 1.1648 π6 + 0.10732 MR8 + 2.6698 R1
- n = 57, r2 = 0.878, s = 0.2779, F = 37.58
Discussion
Conclusions
Appendix
Appendix I. Hydrophobic Fragmental Constant of Groups and Amino Acid Residues
References
- Katritzky, A. A.; Rachwal, P.; Law, K. W.; Karelson, M.; Lobanov, V. S. Prediction of polymer glass transition temperatures using a general quantitative structure-property relationship treatment. J. Chem. Inf. Comput. Sci. 1996, 36, 879–884. [Google Scholar] [CrossRef]
- Bravi, G.; Gancia, E.; Zaliani, A. MS-WHIM, New 3D Theoretical Descriptors Derived from Molecular Surface Properties: A Comparative 3D QSAR Study in A Series of Steroids. J. Computer-aided Mol. Design 1997, 11, 79. [Google Scholar] [CrossRef]
- Estrada, E.; Ramirez, A. Edge Adjacency Relationships and Molecular Topographic Descriptors. Definition and QSAR Applications. J. Chem. Inf. Comput. Sci. 1996, 36, 837. [Google Scholar] [CrossRef]
- Karelson, M.; Lobanov, V.S.; Katritzky, A.R. Quantum-Chemical Descriptors in QSAR/QSPR Studies. Chem Rev. 1996, 96, 1027. [Google Scholar]
- Katrizky, A.P.; Gordeeva, E.V. Traditional Topological Indices vs. Electronic, Geometrical, and Combined Molecular Descriptors in QSAR/QSPR Research. J. Chem. Inf. Comput. Sci. 1993, 33, 835. [Google Scholar] [CrossRef]
- Fujita, M. Organic Analysis; Kakoya bookstore: Tokyo, 1930; p. 33. (in Japanese) [Google Scholar]
- Yuan, L.B.; Ding, Y. Organic Conceptional Diagram and Its Application in Surface Chemistry. Huaxue Tongbao 1988, (2), 19–21. [Google Scholar]
- Wu, H. Study of Group Electronegativities in Biological and Drug Molecules. Zhongguo Kexue Jishu Daxue Xuebao 1990, 20, 517–24. [Google Scholar]
- JavaScript. See http://home.netscape.com/eng/mozilla/3.0/handbook/javascript/index.html.
- Mosley, D.H.; André, J-M. Use of JavaScript in Simple Quantum Chemical Applications. ECCC3. November 1996. at http://hackberry.chem.niu.edu/ECCC3/.
- Rekker, R.F. The Hydrophobic Fragmental Constant, Its Derivation and Application, A Means of Characterizing Membrane Systems; Elsevier: Amsterdam, 1977. [Google Scholar]
- Black, S.D.; Mould, D.R. Development of Hydrophobicity Parameters to Analyze Proteins Which Bear Post- or Cotranslational Modifications. Anal. Biochem. 1991, 193, 72–82. [Google Scholar] [CrossRef]
- Wu, H.; Wen, Y. Computer Aided Anthracycline Anticancer Drugs Design. In 2nd WATOC World Congress, Toronto, 8–14 July 1990.
- Wu, H.; Wen, Y. Quantitative Structure-Activity Relationship Studies of Lincomycin Derivates. In 2nd Symposium on Molecular Mechanics and Drug Design of China, Shanghai, P.R. China, November 1989.
- Wu, H.; Wen, Y. Quantitative Structure-Activity Relationship Studies of Leucomycin and Clindamycin Derivates. In 2nd Symposium on Molecular Mechanics and Drug Design of China, Shanghai, P.R. China, November 1989.
- Wu, H.; Wen, Y. Structure Activity Studies of Podophyllotoxin Derivates. In 2nd Symposium on Molecular Mechanics and Drug Design of China, Shanghai, P.R. China, November 1989.
- Wu, H.; Wen, Y. Structure Activity Studies of Erythromycin Derivates. In 2nd Symposium on Molecular Mechanics and Drug Design of China, Shanghai, P.R. China, November 1989.
- Maddalena, D.J.; Johnston, G.A.R. Prediction of Receptor Properties and Binding Affinity of Ligands to Benzodiazepine/GABAA Receptors Using Articial Neural Networks. J. Med. Chem. 1995, 38, 715–724. [Google Scholar]
- So, S-S.; Karplus, M. Genetic Neural Networks for Quantitative Structure-Activity Relationships: Improvements and Application of Benzodiazepine Affinity for Benzodiazepine/GABAA Receptors. J. Med. Chem. 1996, 39, 5246–5256. [Google Scholar]
- So, S-S.; Karplus, M. Evolution Optimization in Quantitative Structure-Activity Relationship: An Application of Genetic Neural Networks. J. Med. Chem. 1996, 39, 1521–1530. [Google Scholar]
- Gao, H.; Hansch, C. QSAR of P450 Oxidation: On the Value of Comparing Kcat and Km with Kcat/Km. Drug Metabolism Rev. 1996, 28, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Samples Availability: Not available.
| Inorganic group | Value | Organic / inorganic group | Organic Value | Inorganic value |
|---|---|---|---|---|
| -SO2-NH- | 240 | -OSO3H | 20 | 220 |
| =N-OH | 210 | >SO2 | 40 | 170 |
| -NHCONH- | ||||
| -CO-NH-* | 200 | >SO | 40 | 140 |
| -COOH | 150 | -SCN | 90 | 80 |
| R-C|H(CH2)n-C=OO| | 120 | -NCS | 90 | 75 |
| -COO- (in a ring above) | ||||
| -CO-O-CO- | 110 | -NO2 | 70 | 70 |
| Anthracene or Phenanthrene nuclei | 105 | -As< | 40 | 70 |
| -CN | ||||
| -OH | 100 | -P< | 20 | 70 |
| >Hg (covalent) | 95 | -O- [CH2CH2O] -CH2- | 30 | 60 |
| -NH-NH- | 80 | -NO | 50 | 50 |
| -O-CO-O- | ||||
| -N< (-NH2,-NHPh,-NPh2) | 70 | -O-NO2 | 60 | 40 |
| >CO | 65 | -NC | 40 | 40 |
| -COOPh | 60 | -P=P- | 30 | 30 |
| naphthalene nuclei | -NCO | |||
| >C=NH | 50 | -ONO | 40 | 20 |
| -SH | ||||
| -S- | ||||
| -O-O- | 40 | -I | 80 | 10 |
| -N=N- | 30 | -Br | 60 | 10 |
| -O- | 20 | =S | 50 | 10 |
| Benzene nuclei | 15 | -Cl | 40 | 10 |
| (single aromatic ring) | ||||
| Ring (non aromatic single ring) | 10 | -F | 5 | 5 |
| Triple bond | 3 | Iso-** | −10 | 0 |
| Double bond | 2 | Tert-** | −20 | 0 |
| Group | π | Mr | F | R | σm | σp | I | O | X |
|---|---|---|---|---|---|---|---|---|---|
| F | 0.14 | 0.92 | 0.43 | −0.34 | 0.34 | 0.06 | 5 | 5 | 3.98 |
| Cl | 0.71 | 6.03 | 0.41 | −0.15 | 0.37 | 0.23 | 10 | 40 | 3.16 |
| Br | 0.86 | 8.88 | 0.44 | −0.17 | 0.39 | 0.23 | 10 | 50 | 2.96 |
| I | 1.12 | 13.94 | 0.4 | −0.19 | 0.35 | 0.18 | 10 | 60 | 2.66 |
| H | 0 | 1.03 | 0 | 0 | 0 | 0 | 0 | 0 | 2.20 |
| OH | −0.67 | 2.85 | 0.29 | −0.64 | 0.12 | −0.37 | 100 | 0 | 3.03 |
| SH | 0.39 | 9.22 | 0.28 | −0.11 | 0.25 | 0.15 | 20 | 40 | 2.45 |
| NH2 | −1.23 | 5.42 | 0.02 | −0.68 | −0.16 | −0.66 | 70 | 0 | 2.70 |
| NHOH | −1.34 | 7.22 | 0.06 | −0.4 | −0.04 | −0.34 | 170 | 0 | 2.87 |
| NHNH2 | −0.88 | 8.44 | 0.17 | −0.71 | −0.02 | −0.55 | 140 | 0 | 2.80 |
| CF3 | 0.88 | 5.02 | 0.38 | 0.19 | 0.43 | 0.54 | 15 | 35 | 3.16 |
| OCF3 | 1.04 | 7.86 | 0.38 | 0 | 0.38 | 0.35 | 35 | 35 | 3.35 |
| CN | −0.57 | 6.33 | 0.51 | 0.19 | 0.56 | 0.66 | 70 | 40 | 2.76 |
| NCS | 1.15 | 17.24 | 0.51 | −0.09 | 0.48 | 0.38 | 75 | 90 | 2.85 |
| SCN | 0.41 | 13.4 | 0.36 | 0.19 | 0.41 | 0.52 | 80 | 90 | 2.64 |
| NHCN | −0.26 | 10.14 | 0.26 | −0.18 | 0.21 | 0.06 | 140 | 40 | 2.82 |
| CHO | −0.65 | 6.88 | 0.31 | 0.13 | 0.35 | 0.42 | 65 | 20 | 2.75 |
| CO2H | −0.32 | 6.93 | 0.33 | 0.15 | 0.37 | 0.45 | 150 | 20 | 2.87 |
| CH2Br | 0.79 | 13.39 | 0.1 | 0.05 | 0.12 | 0.14 | 10 | 80 | 2.51 |
| CH2Cl | 0.17 | 10.49 | 0.1 | 0.03 | 0.11 | 0.12 | 10 | 60 | 2.54 |
| CH2I | 1.5 | 18.6 | 0.09 | 0.03 | 0.1 | 0.11 | 10 | 100 | 2.47 |
| NHCHO | −0.98 | 10.31 | 0.25 | −0.23 | 0.19 | 0 | 135 | 20 | 2.81 |
| CONH2 | −1.49 | 9.81 | 0.24 | 0.14 | 0.28 | 0.36 | 135 | 20 | 2.83 |
| CH=NOH | −0.38 | 10.28 | 0.25 | −0.13 | 0.22 | 0.1 | 220 | 20 | 2.64 |
| CH3 | 0.56 | 5.65 | −0.04 | −0.13 | −0.07 | −0.17 | 0 | 20 | 2.40 |
| NHCONH2 | −1.3 | 13.72 | 0.04 | −0.28 | −0.03 | −0.24 | 105 | 20 | 2.83 |
| OCH3 | −0.02 | 7.87 | 0.26 | −0.51 | 0.12 | −0.27 | 20 | 20 | 3.09 |
| CH2OH | −1.03 | 7.19 | 0 | 0 | 0 | 0 | 100 | 20 | 2.52 |
| NHCH3 | −0.47 | 10.33 | −0.11 | −0.74 | −0.3 | −0.84 | 70 | 20 | 2.74 |
| -C°CH | 0.4 | 9.55 | 0.19 | 0.05 | 0.21 | 0.23 | 3 | 40 | 2.50 |
| NHCOCF3 | 0.08 | 14.3 | 0.36 | −0.21 | 0.3 | 0.12 | 150 | 40 | 2.84 |
| CH2CN | −0.57 | 10.11 | 0.21 | −0.18 | 0.16 | 0 | 70 | 60 | 2.48 |
| -CH=CH2 | 0.82 | 10.99 | 0.07 | −0.08 | 0.05 | −0.02 | 2 | 40 | 2.46 |
| COCH3 | −0.55 | 11.18 | 0.32 | 0.2 | 0.38 | 0.5 | 65 | 40 | 2.78 |
| OCOCH3 | −0.64 | 12.47 | 0.41 | −0.07 | 0.39 | 0.31 | 85 | 40 | 3.22 |
| CO2CH3 | −0.01 | 12.87 | 0.33 | 0.15 | 0.37 | 0.45 | 85 | 40 | 2.88 |
| NHCOCH3 | −0.97 | 14.93 | 0.28 | −0.26 | 0.21 | 0 | 135 | 40 | 2.82 |
| NHCO2CH3 | −0.37 | 16.53 | 0.14 | −0.28 | 0.07 | −0.15 | 155 | 40 | 2.84 |
| CH2CH3 | 1.02 | 10.3 | −0.05 | −0.1 | −0.07 | −0.15 | 0 | 40 | 2.43 |
| CH2OCH3 | −0.78 | 12.07 | 0.01 | 0.02 | 0.02 | 0.03 | 20 | 40 | 2.53 |
| OCH2CH3 | 0.38 | 12.47 | 0.22 | −0.44 | 0.1 | −0.24 | 20 | 40 | 3.10 |
| N(CH3)2 | 0.18 | 15.55 | 0.1 | −0.92 | −0.15 | −0.83 | 70 | 40 | 2.78 |
| CH(CH3)2 | 1.53 | 14.96 | −0.05 | −0.1 | −0.07 | −0.15 | 0 | 50 | 2.46 |
| C3H7 | 1.55 | 14.96 | −0.06 | −0.08 | −0.07 | −0.13 | 0 | 60 | 2.43 |
| C4H9 | 2.13 | 19.61 | −0.06 | −0.11 | −0.08 | −0.16 | 0 | 80 | 2.43 |
| C(CH3)3 | 1.98 | 19.62 | −0.07 | −0.13 | −0.1 | −0.2 | 0 | 60 | 2.49 |
| C6H5 | 1.96 | 25.36 | 0.08 | −0.08 | 0.06 | −0.01 | 15 | 120 | 2.51 |
| C5H11 | 2.67 | 24.26 | −0.06 | −0.08 | −0.08 | −0.16 | 0 | 100 | 2.43 |
| OC6H5 | 2.08 | 27.68 | 0.34 | −0.35 | 0.25 | −0.03 | 35 | 120 | 3.13 |
| N(C2H5)2 | 1.18 | 24.85 | 0.01 | −0.91 | −0.23 | −0.9 | 70 | 80 | 2.80 |
| Descriptors | π | Mr | F | R | σm | σp | I | O |
|---|---|---|---|---|---|---|---|---|
| Mr | 0.601 | |||||||
| F | −0.175 | −0.251 | ||||||
| R | 0.116 | −0.104 | 0.243 | |||||
| σm | −0.105 | −0.250 | 0.934 | 0.574 | ||||
| σp | 0.007 | −0.200 | 0.650 | 0.895 | 0.879 | |||
| I | −0.667 | −0.109 | 0.234 | −0.171 | 0.136 | −0.025 | ||
| O | 0.752 | 0.837 | −0.030 | 0.206 | 0.050 | 0.144 | −0.378 | |
| X | −0.193 | −0.245 | 0.637 | −0.219 | 0.459 | 0.122 | 0.186 | −0.244 |
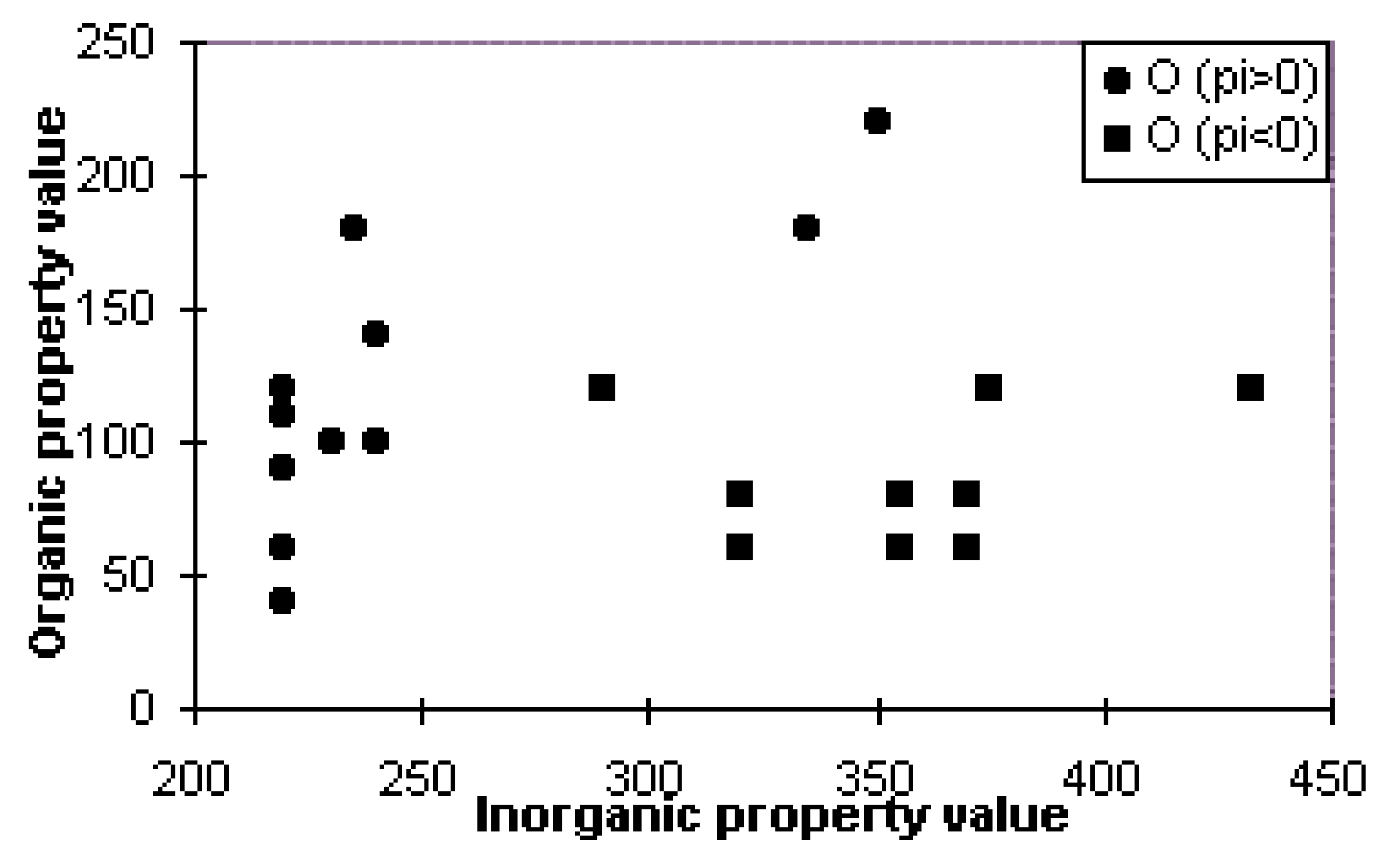
| Amino acid(residue) | logP Value | Inorganic (I) Value | Organic (O) Value | Residue Electronegativity (X) |
|---|---|---|---|---|
| A (Ala) | 0.072 | 220 | 60 | 2.40 |
| R (Arg) | −2.061 | 432 | 120 | 2.43 |
| N (Asn) | −1.003 | 355 | 60 | 2.49 |
| D (Asp) | −1.935 | 370 | 60 | 2.50 |
| C (Cys) | 0.987 | 240 | 100 | 2.44 |
| Q (Gln) | −0.936 | 355 | 80 | 2.49 |
| E (Glu) | −1.868 | 370 | 80 | 2.44 |
| G (Gly) | 0.184 | 220 | 40 | 2.20 |
| H (His) | −1.321 | 375 | 120 | 2.45 |
| I (Ile) | 2.167 | 220 | 120 | 2.46 |
| L (Leu) | 2.167 | 220 | 110 | 2.44 |
| K (Lys) | −0.790 | 290 | 120 | 2.43 |
| M (Met) | 1.246 | 240 | 140 | 2.44 |
| F (Phe) | 2.423 | 235 | 180 | 2.45 |
| P (Pro) | 1.128 | 230 | 100 | / |
| S (Ser) | −0.453 | 320 | 60 | 2.52 |
| T (Thr) | −0.042 | 320 | 80 | 2.45 |
| W (Trp) | 1.878 | 335 | 220 | 2.45 |
| Y (Tyr) | 1.887 | 335 | 180 | 2.45 |
| V (Val) | 1.640 | 220 | 90 | 2.46 |
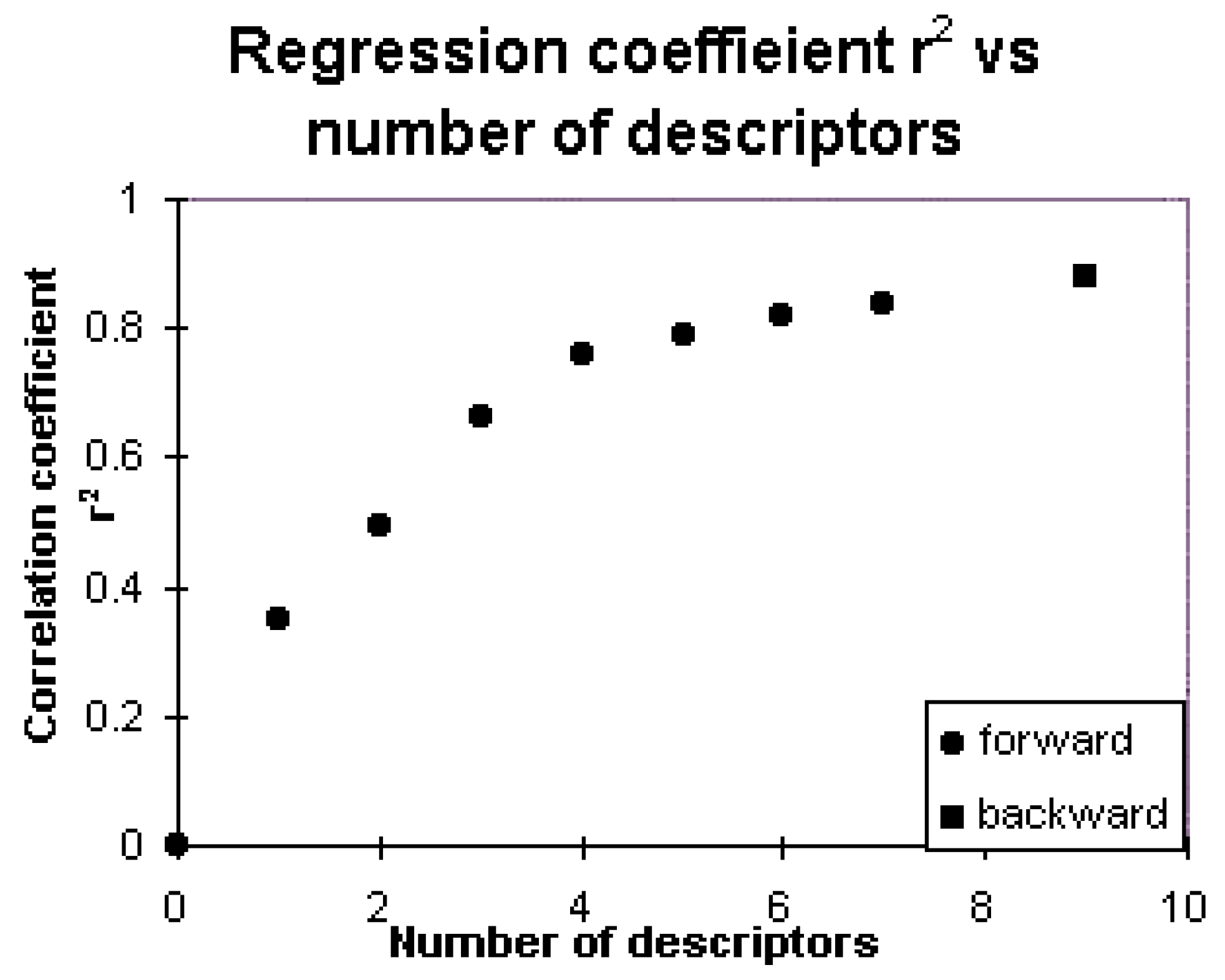
| Step | 1 | 2 | 3 | 4 | 5 | 6 | 7* |
| Constant | 1.757 | 1.444 | 1.506 | 1.850 | 2.098 | 2.145 | 1.998 |
| σm2 | −2.54 | −2.31 | −1.91 | −1.78 | −1.79 | −1.64 | −1.68 |
| T-ratio | −5.44 | −5.50 | −5.36 | −5.84 | −6.20 | −6.03 | −6.42 |
| MR1 | 0.067 | 0.093 | 0.106 | 0.100 | 0.126 | 0.131 | |
| T-ratio | 3.97 | 6.25 | 8.15 | 7.93 | 8.72 | 9.33 | |
| π (total) | −0.363 | −0.352 | −0.234 | −0.221 | −0.290 | ||
| T-ratio | −5.08 | −5.78 | −3.23 | −3.29 | −4.07 | ||
| F7 | −1.15 | −1.71 | −1.85 | −1.76 | |||
| T-ratio | −4.55 | −5.38 | −6.19 | −6.09 | |||
| π7 | −0.209 | −0.239 | −0.203 | ||||
| T-ratio | −2.69 | −3.29 | −2.85 | ||||
| R1 | 2.33 | 2.36 | |||||
| T-ratio | 3.06 | 3.22 | |||||
| MR6 | 0.116 | ||||||
| T-ratio | 2.30 | ||||||
| s | 0.593 | 0.527 | 0.436 | 0.372 | 0.352 | 0.326 | 0.313 |
| r2 | 0.3496 | 0.4968 | 0.6615 | 0.7578 | 0.7879 | 0.8213 | 0.8387 |
| F | 29.56 | 26.65 | 34.53 | 40.68 | 37.89 | 38.29 | 36.39 |
© 1999 MDPI. All rights reserved.
Share and Cite
Wu, H. Chemical Property Calculation through JavaScript and Applications in QSAR. Molecules 1999, 4, 16-27. https://doi.org/10.3390/40100016
Wu H. Chemical Property Calculation through JavaScript and Applications in QSAR. Molecules. 1999; 4(1):16-27. https://doi.org/10.3390/40100016
Chicago/Turabian StyleWu, Hanqing. 1999. "Chemical Property Calculation through JavaScript and Applications in QSAR" Molecules 4, no. 1: 16-27. https://doi.org/10.3390/40100016




