Bile Acids as Building Blocks of Supramolecular Hosts
Abstract
:1. Introduction
2. Structure, Metabolism, and Functions of Bile Acids

3. Pharmacological Applications of Bile Acids and Their Derivatives
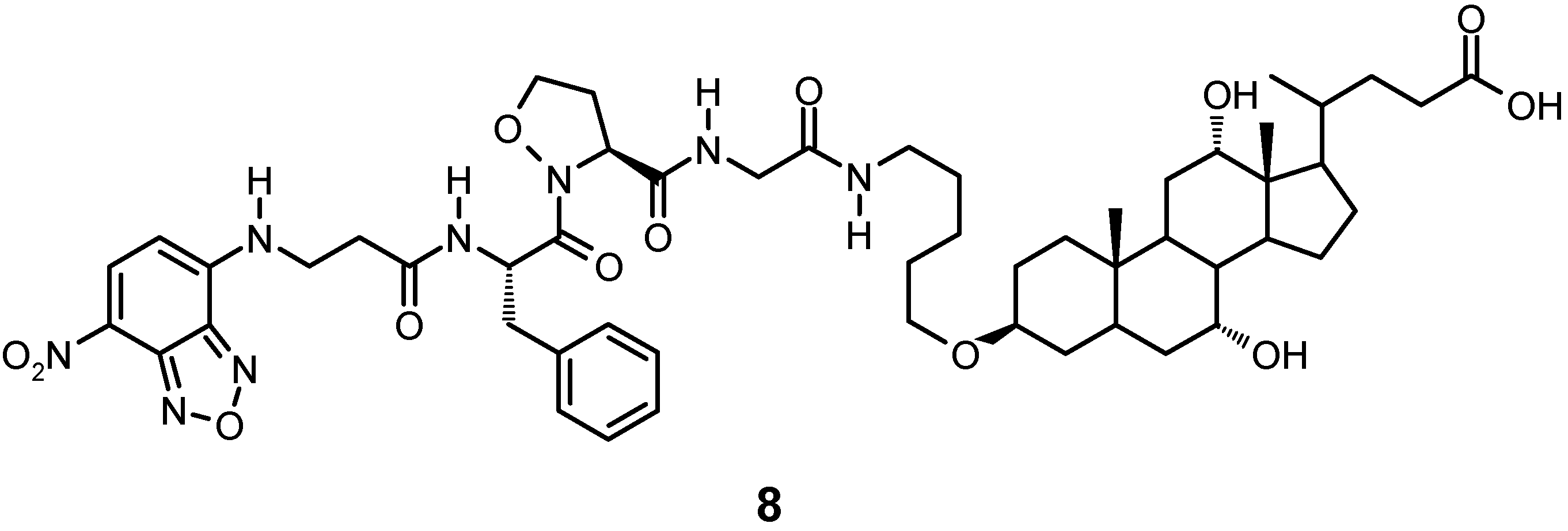
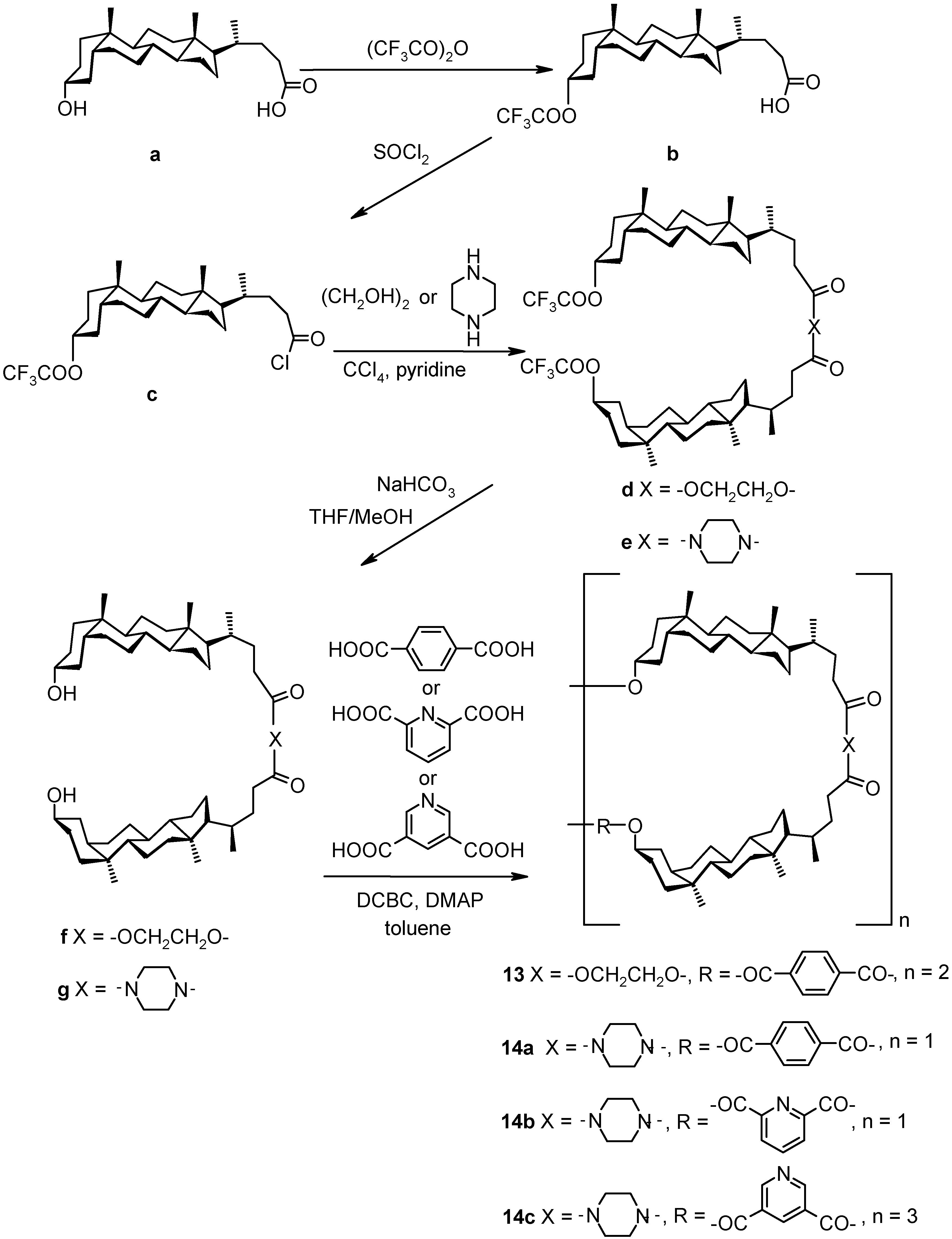
4. Bile Acid-based Molecular and Supramolecular Assemblies
4.1. Cyclic Compounds
4.1.1. Cyclocholates
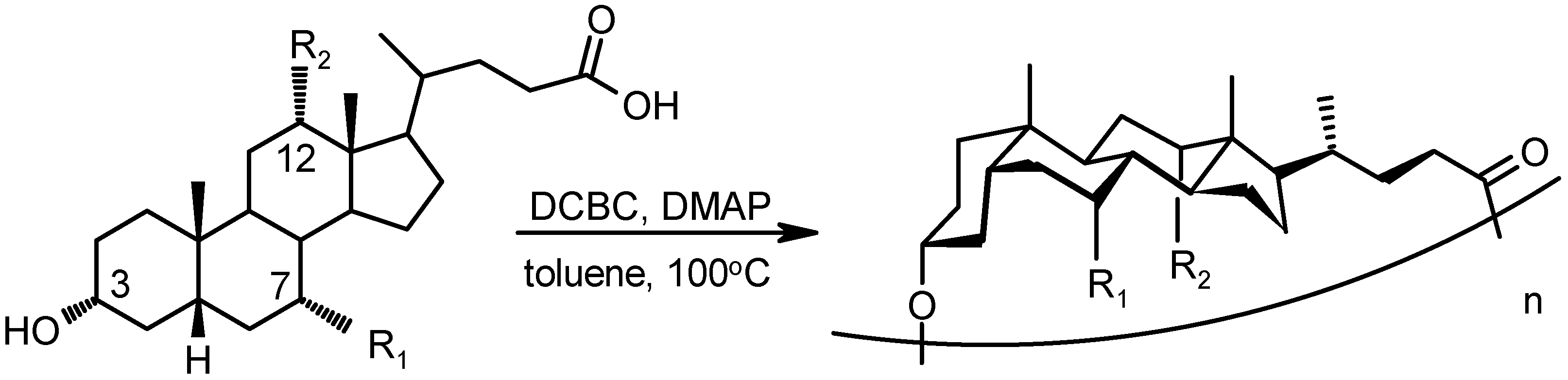


4.1.2. Cholaphanes
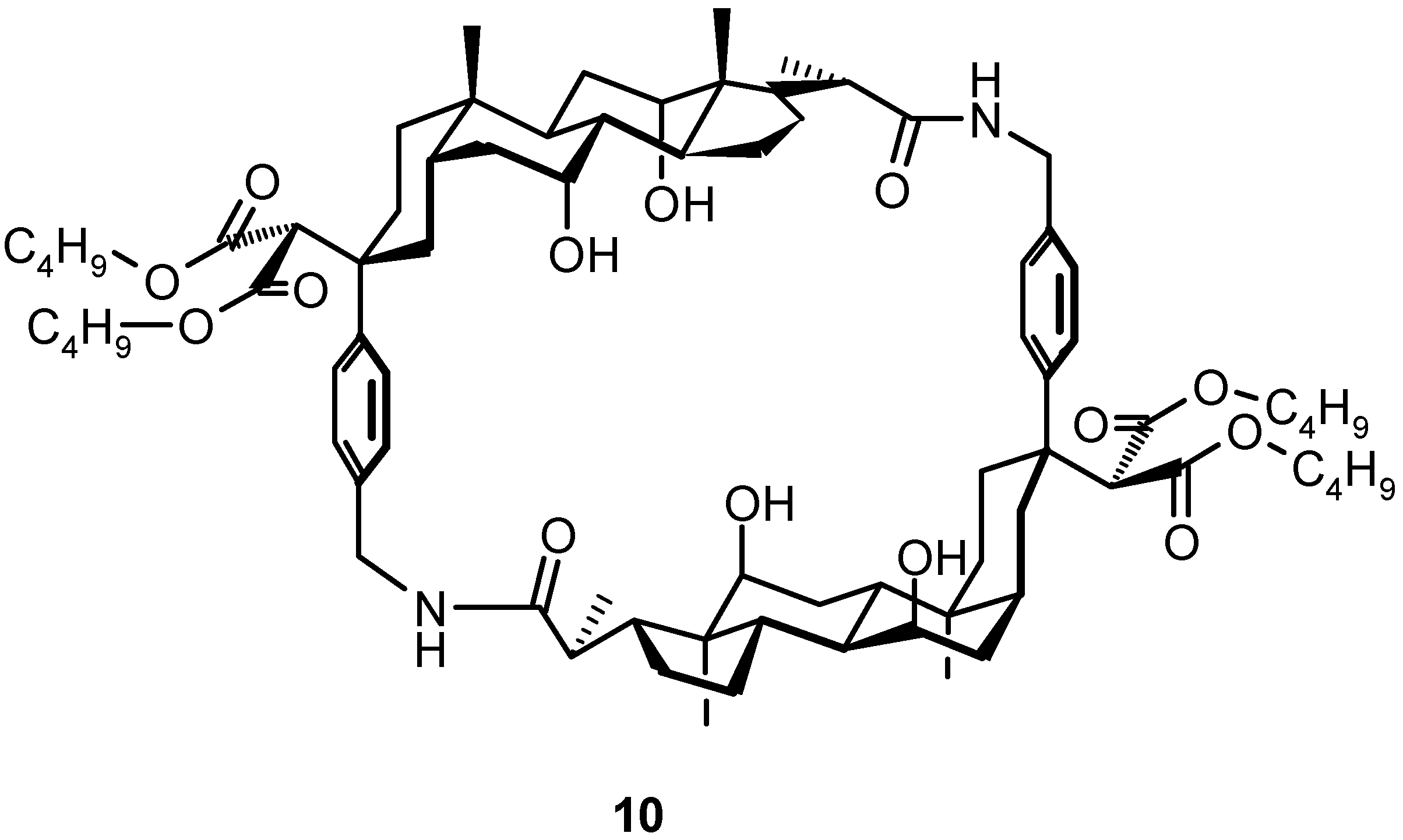
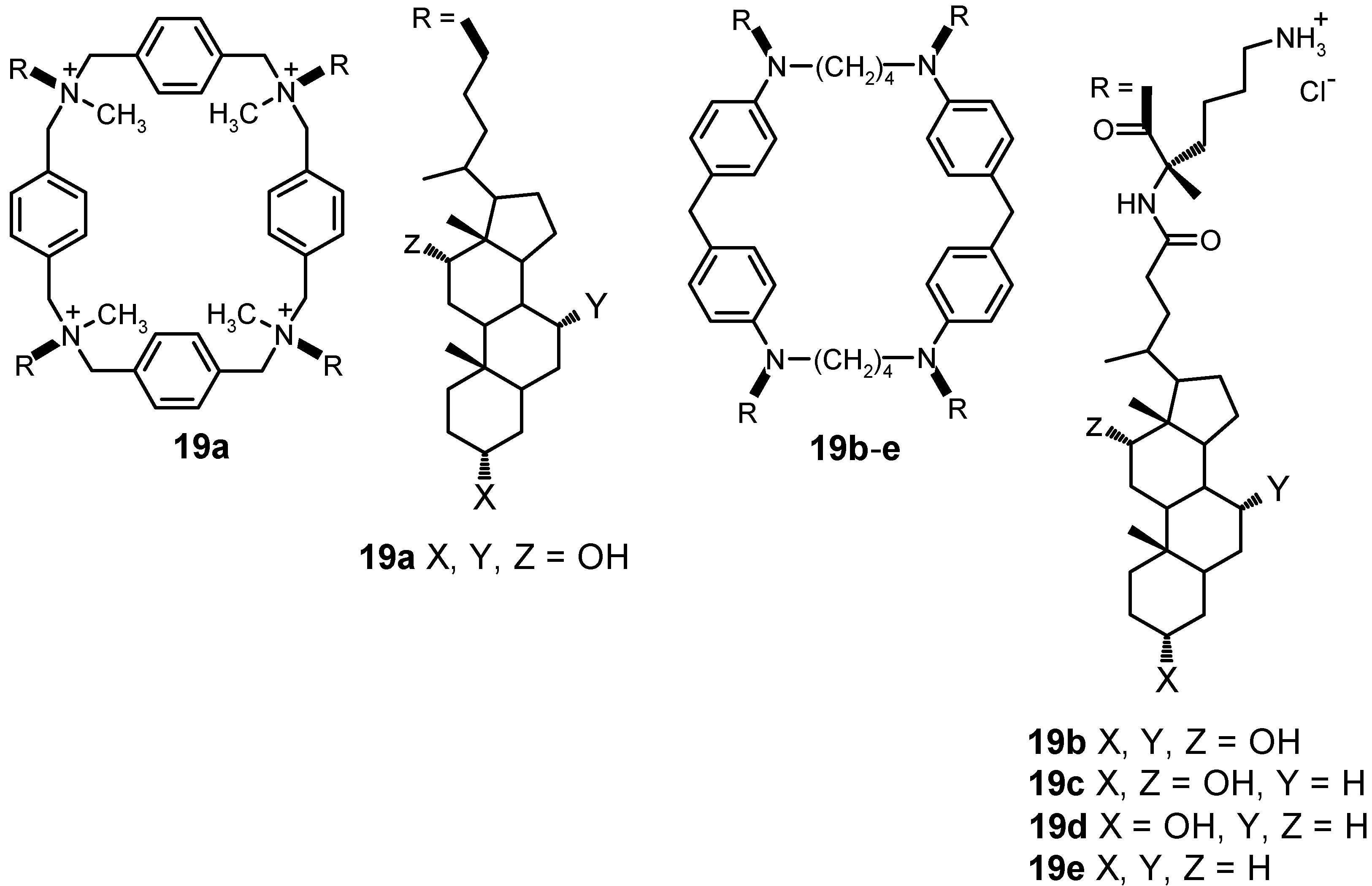
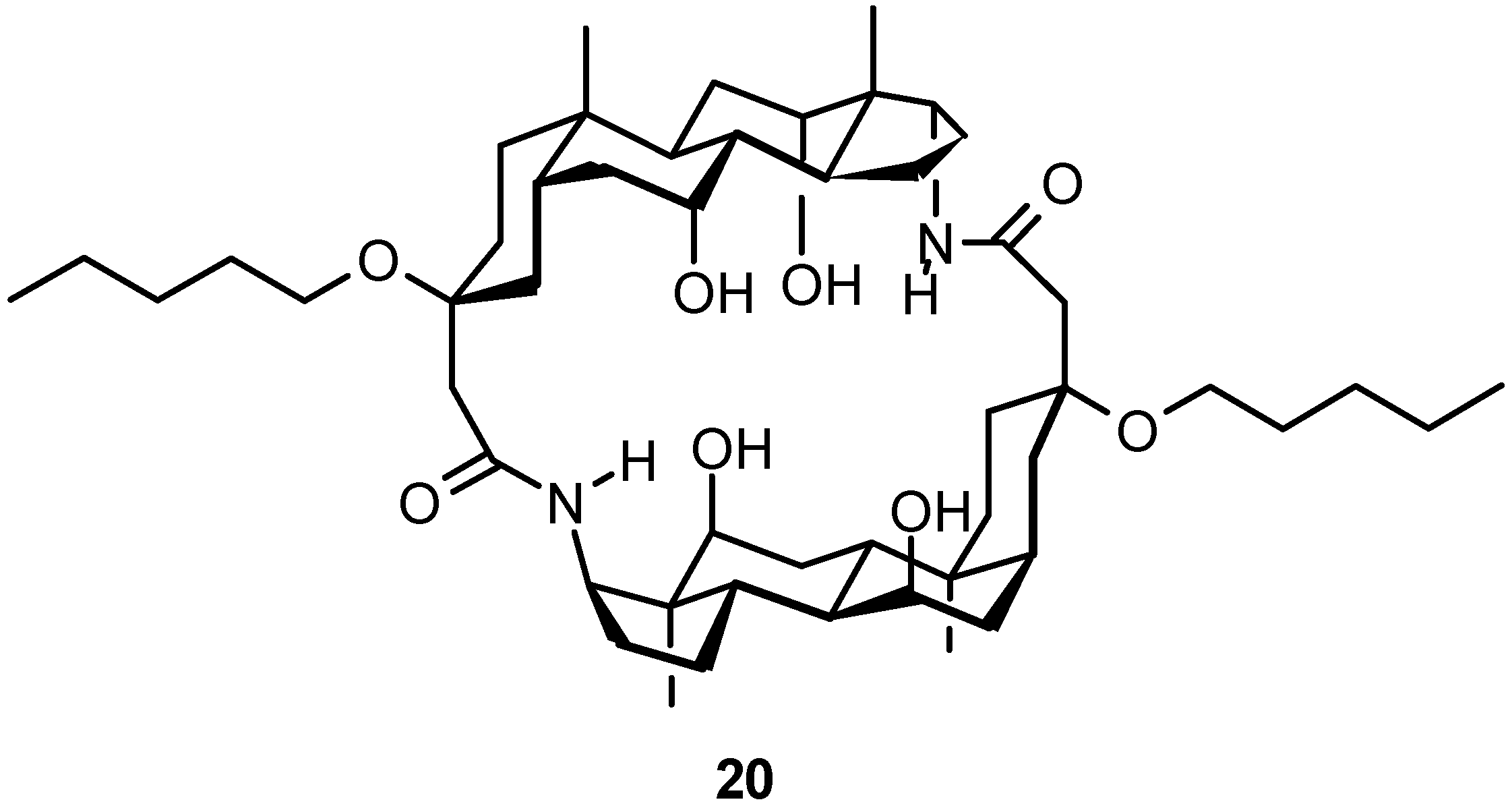
4.1.3. Other Cyclic Structures
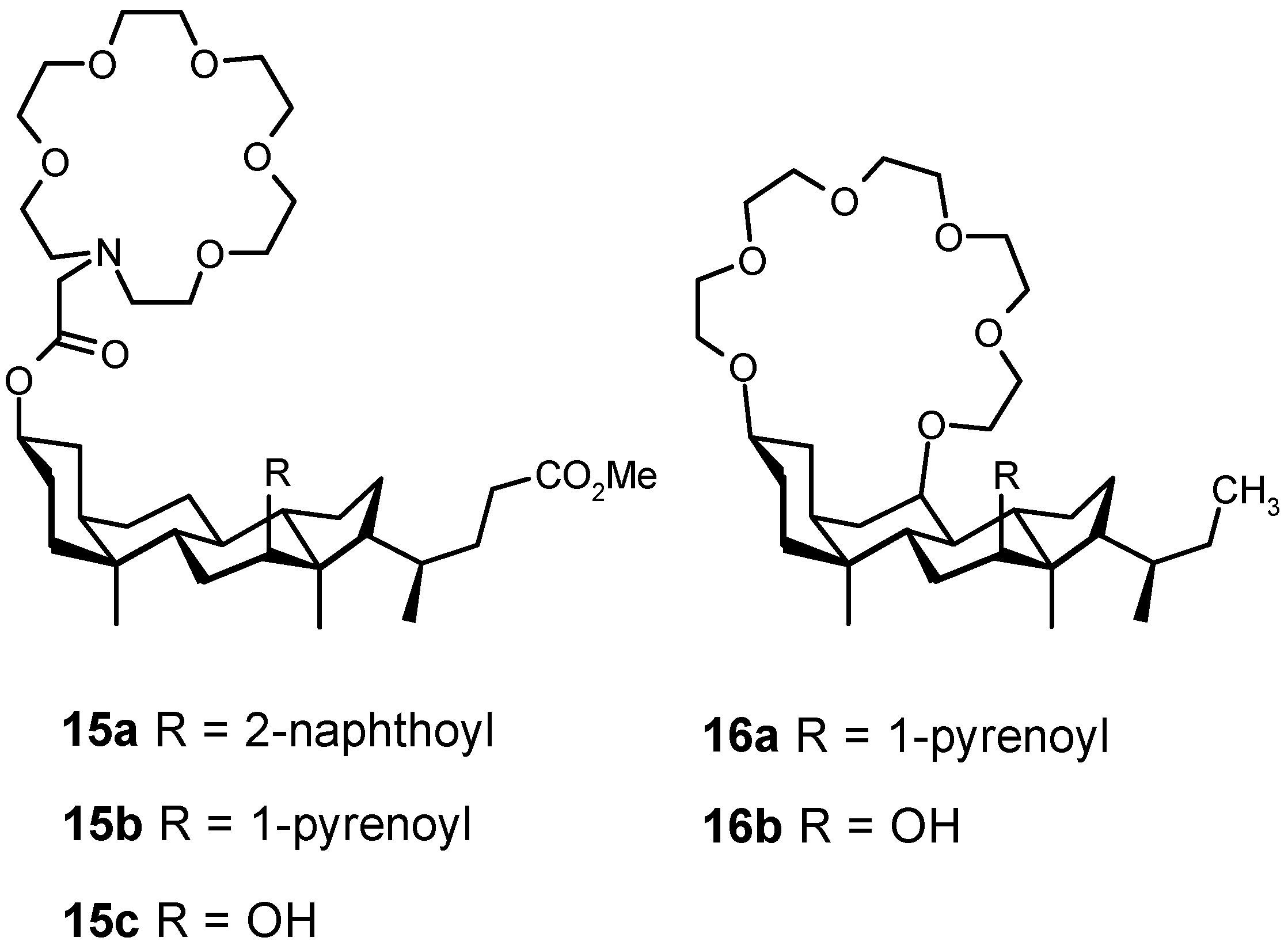

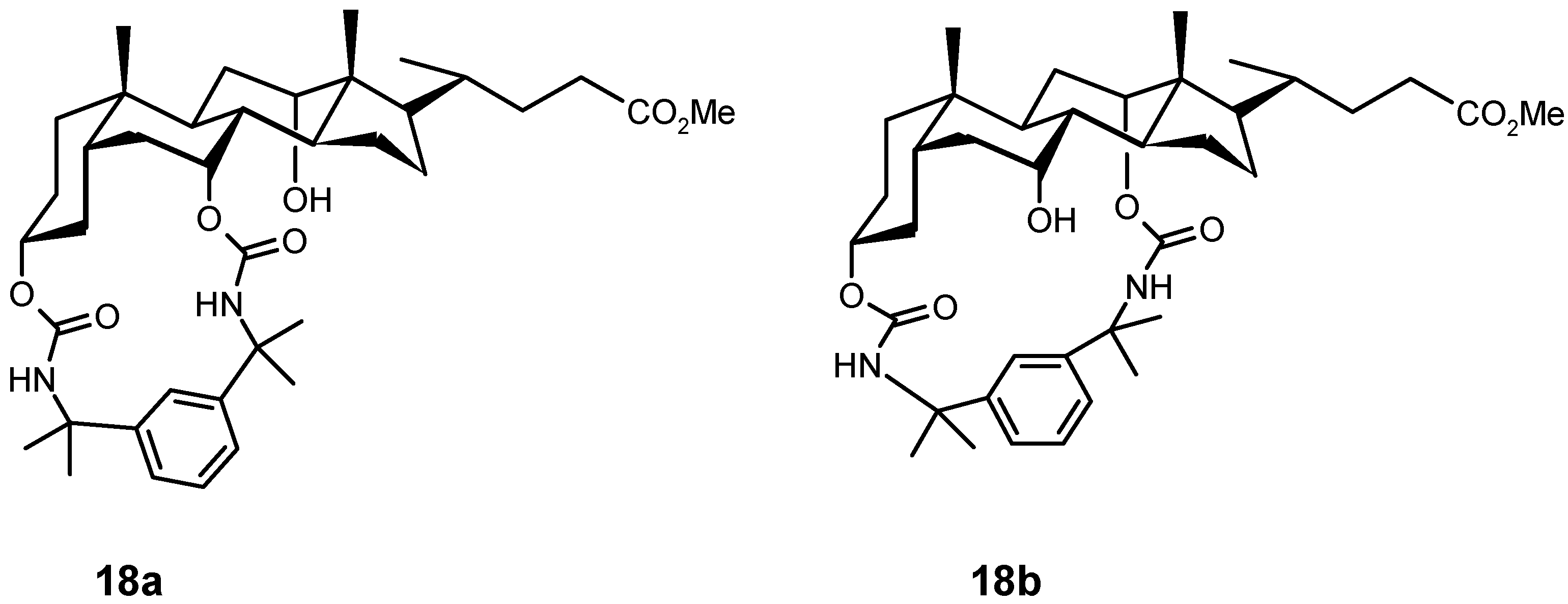
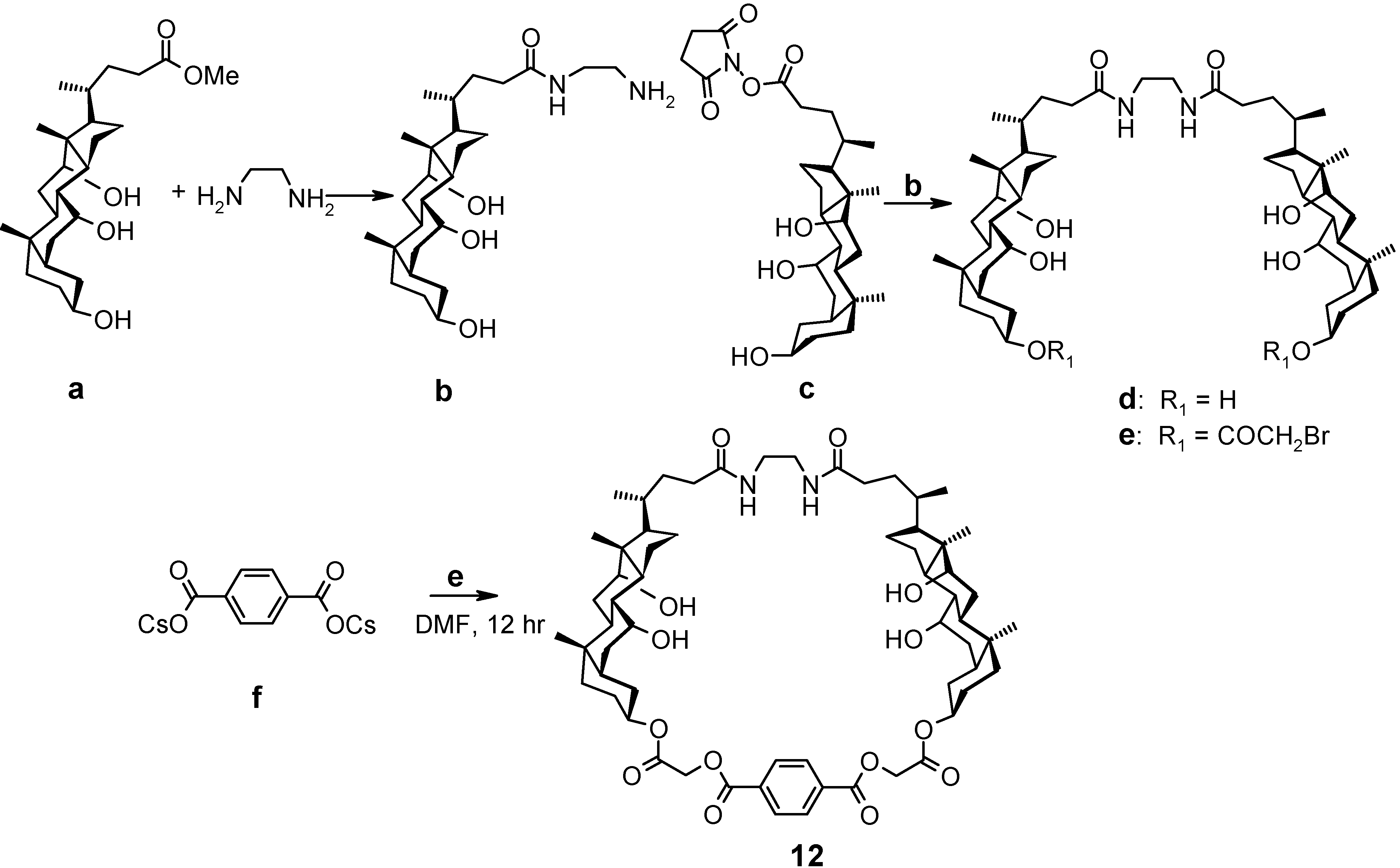
4.2. Acyclic Compounds
4.2.1. Cleft-type Structures
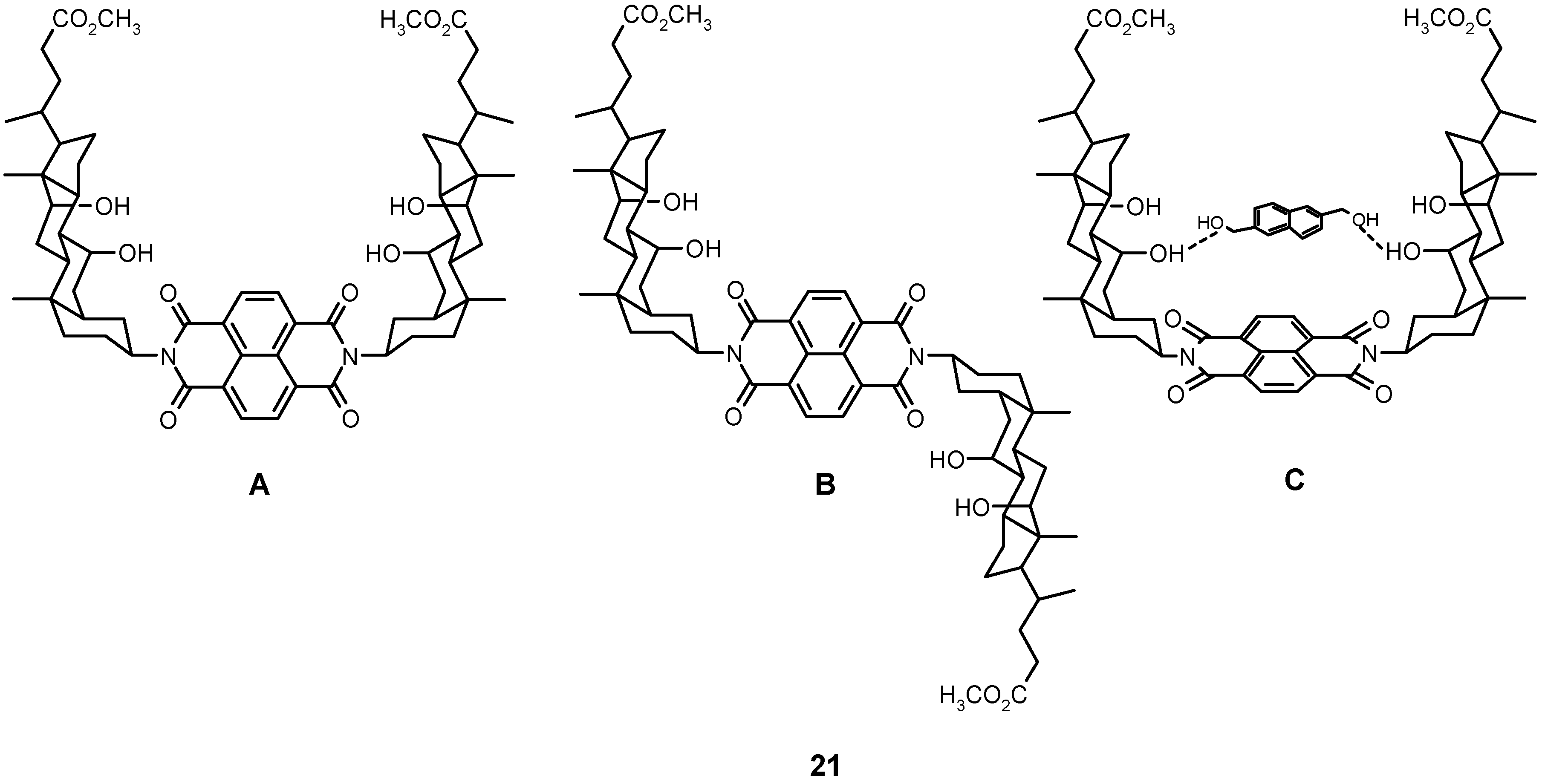
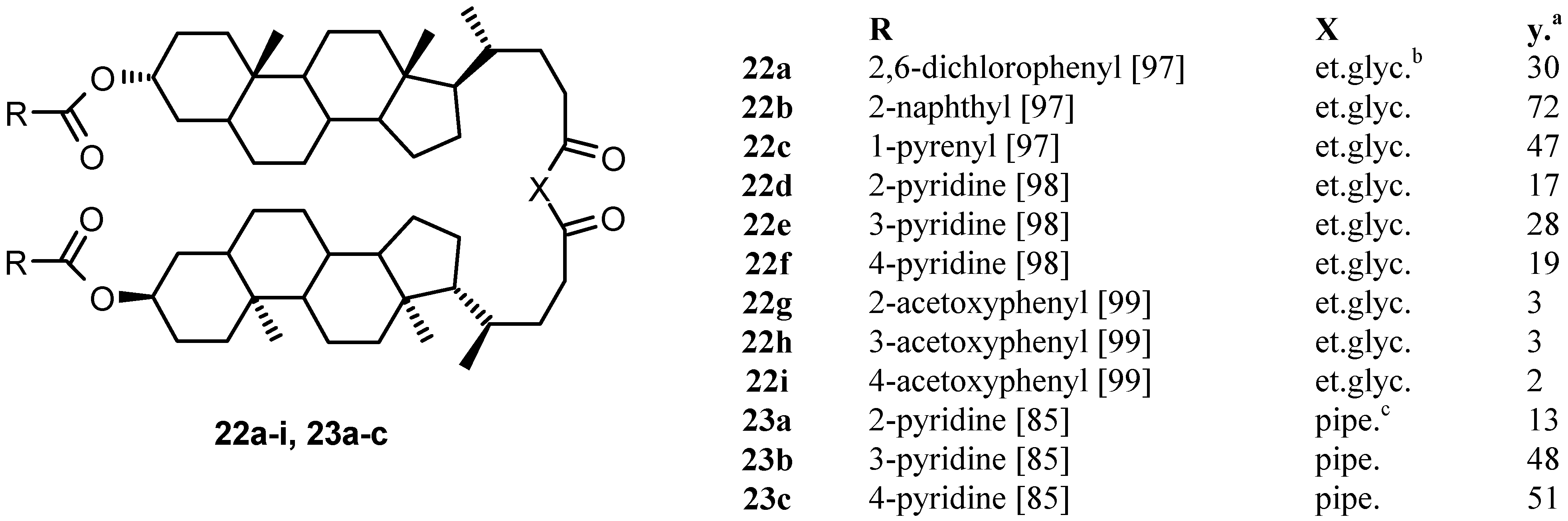
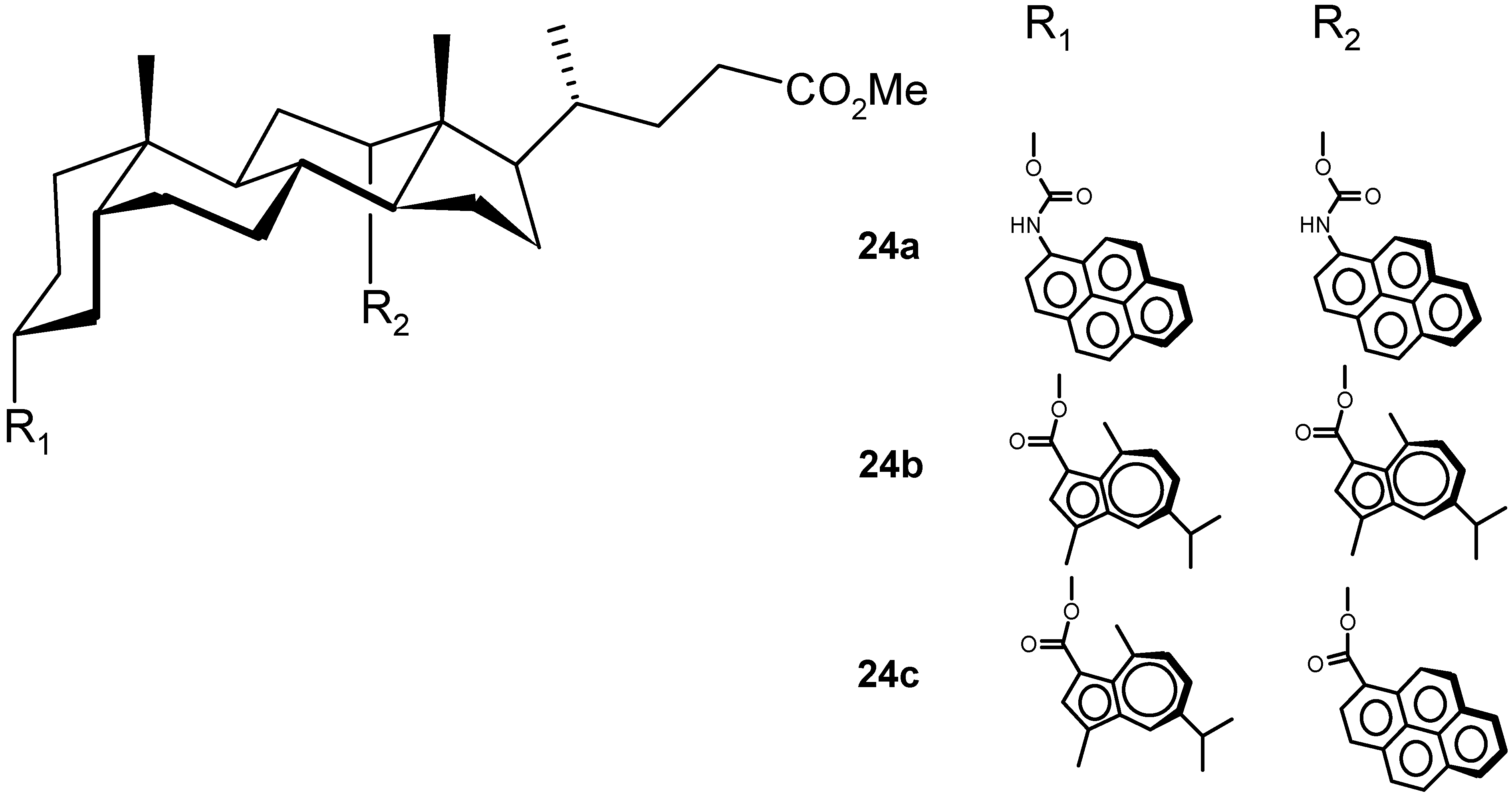
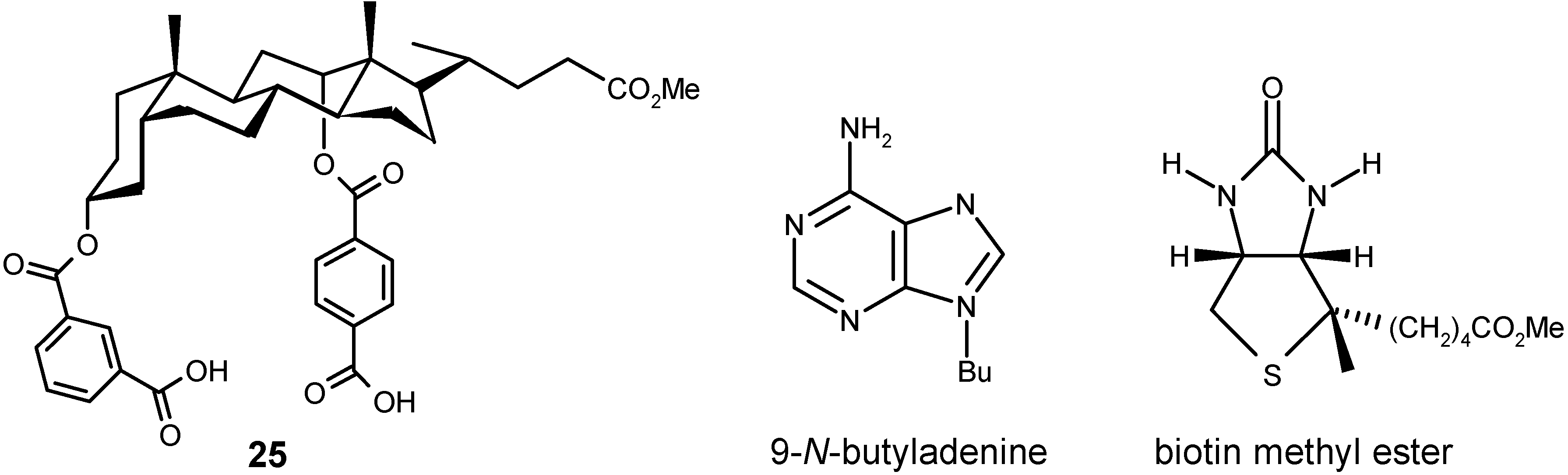
4.2.2. Other Acyclic Structures
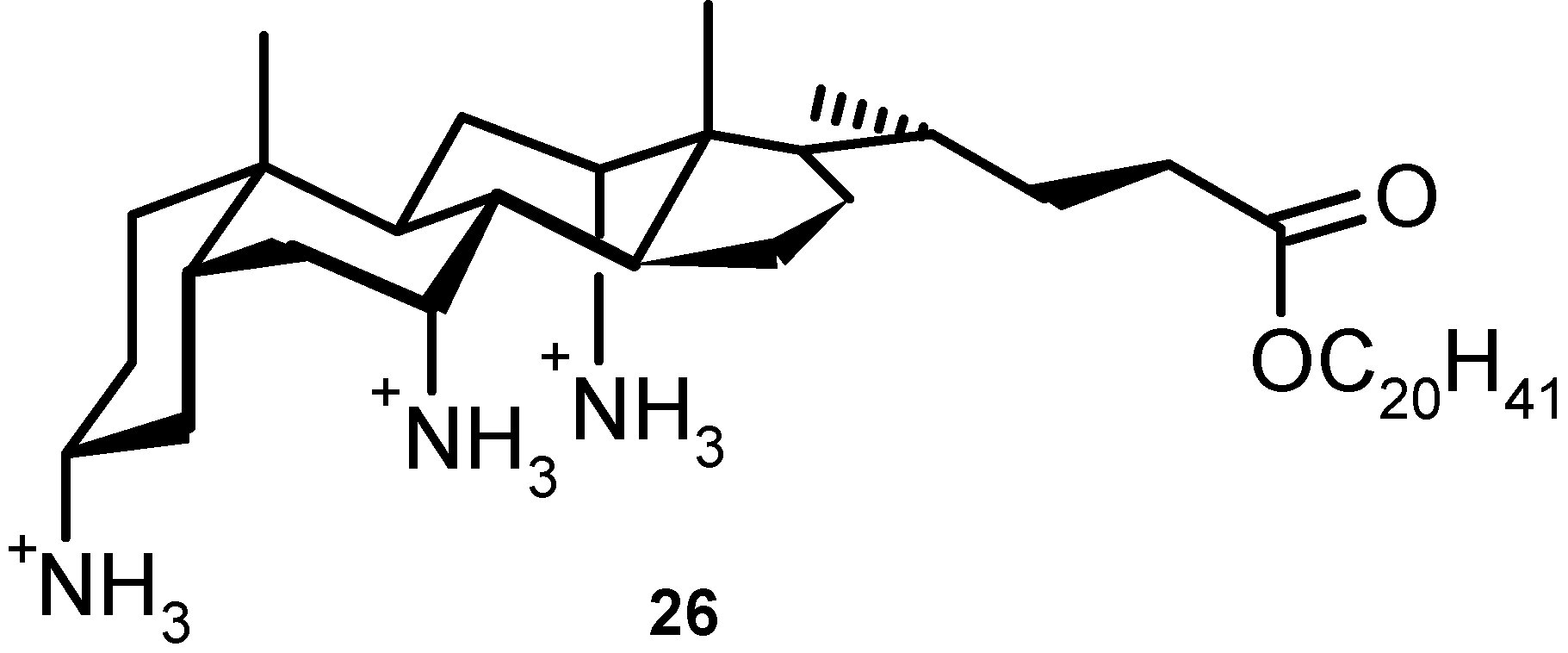
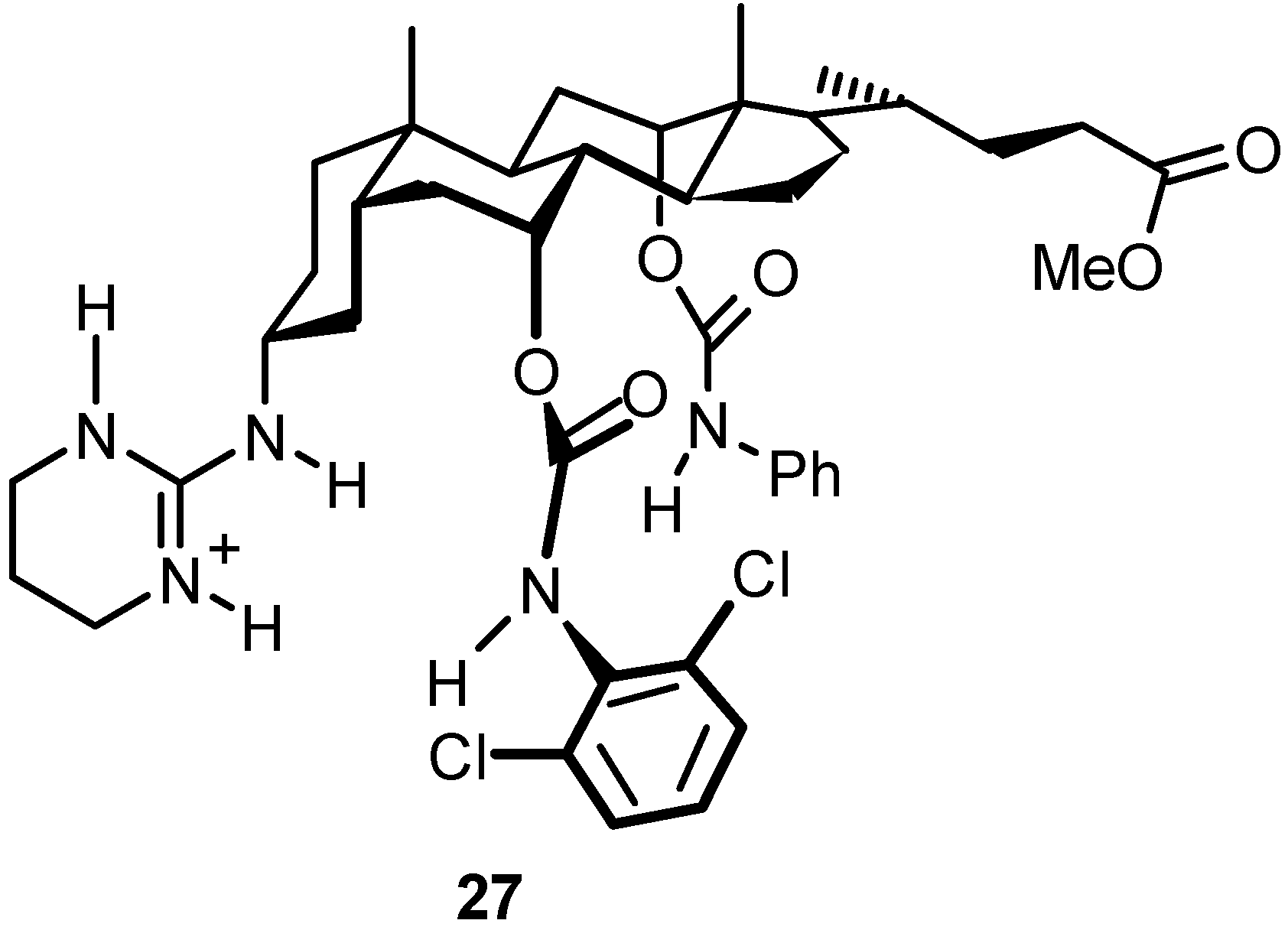
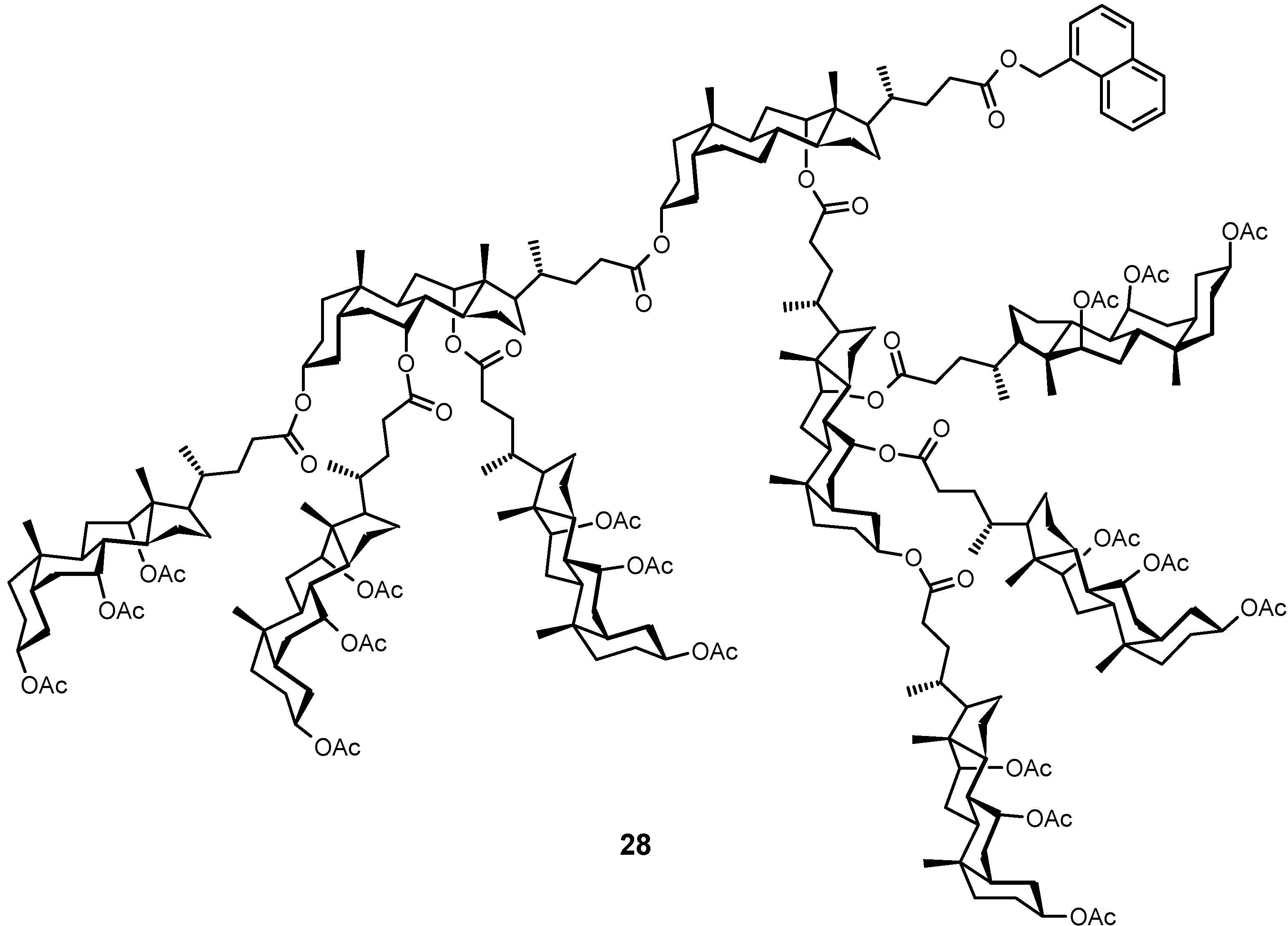
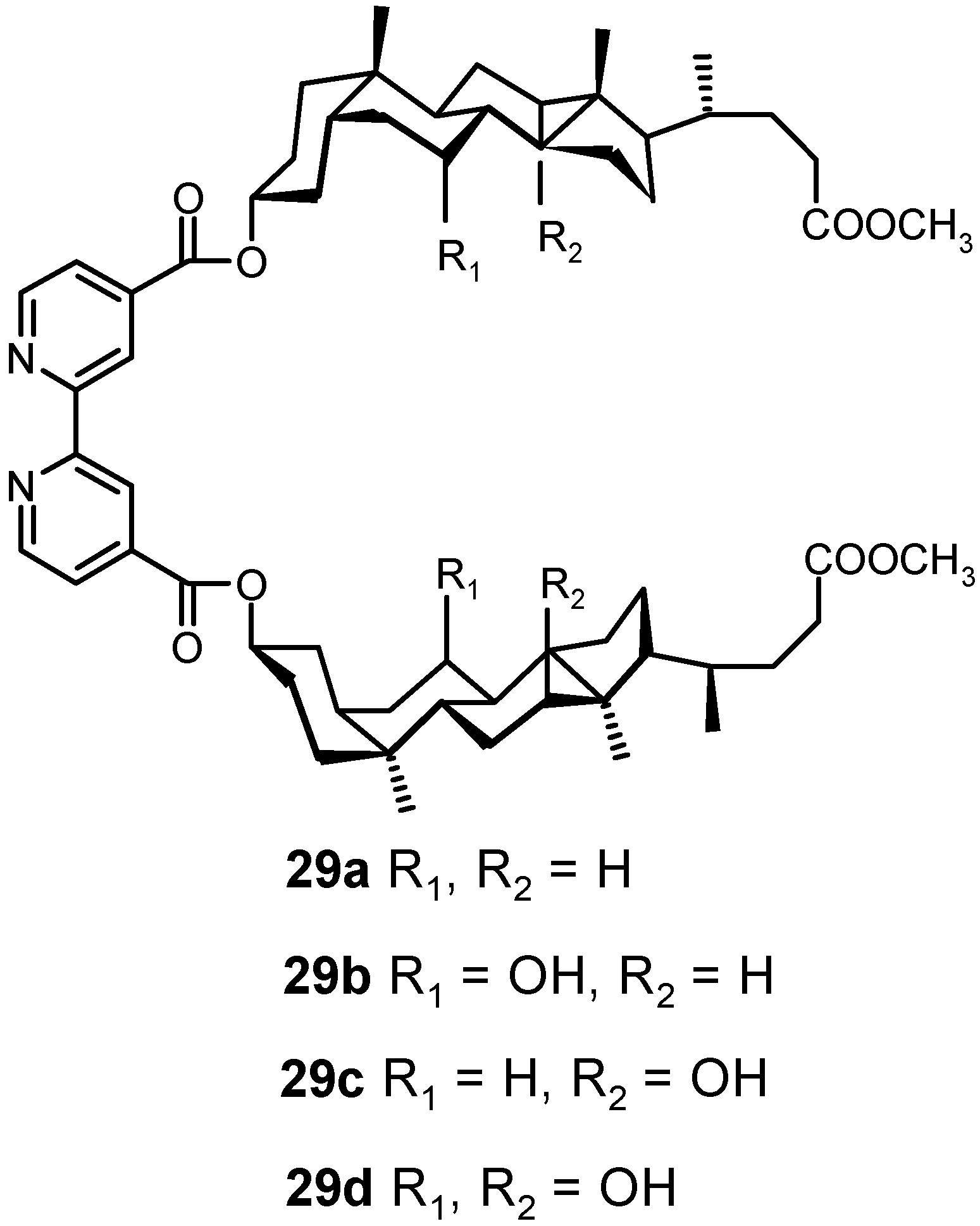
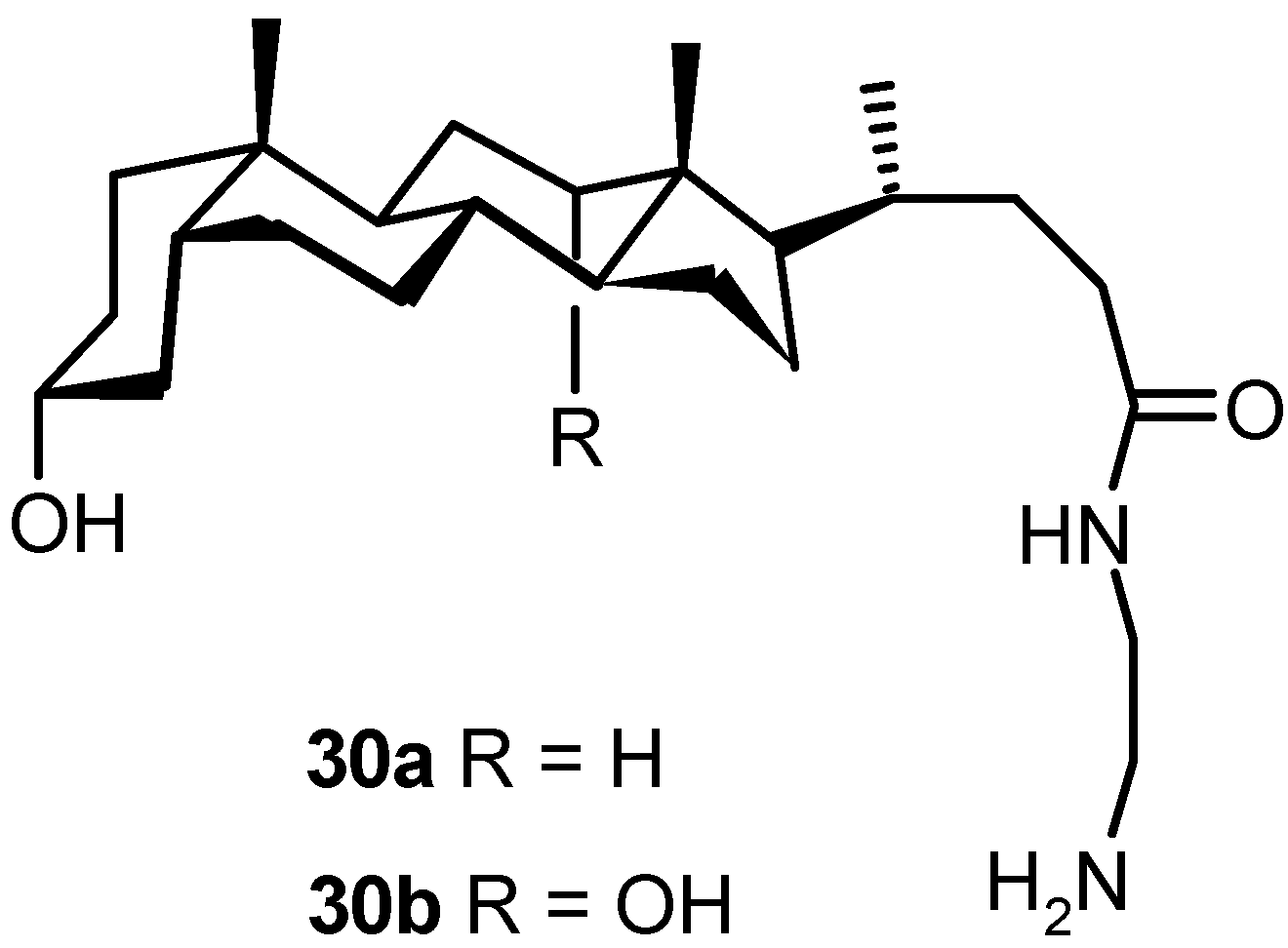
4.3. Inclusion Complexes
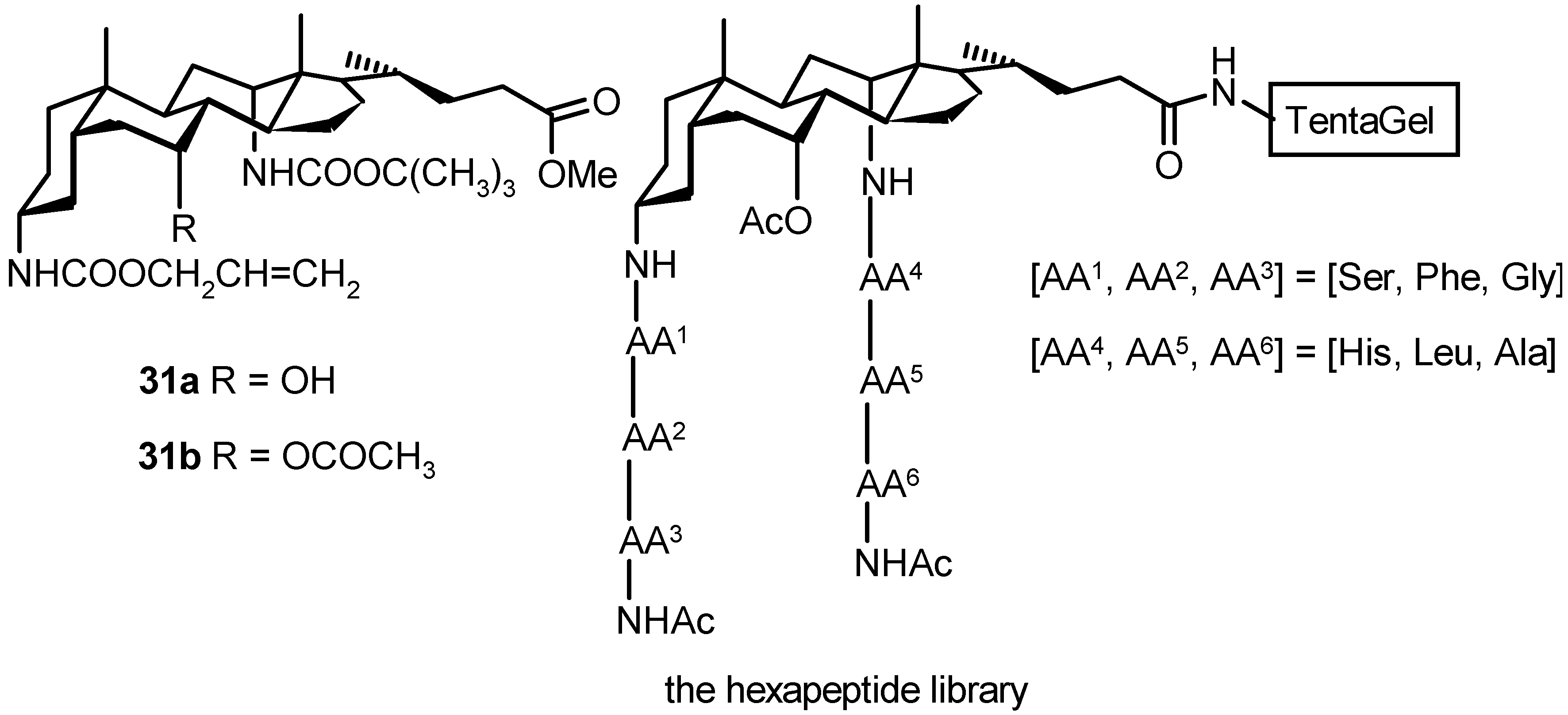
4.4. Bile Acids in Combinatorial Chemistry
5. The Latest Spectroscopic and Computational Studies of Bile Acids and Their Derivatives
5.1. NMR Spectroscopy
5.2. Mass Spectroscopy
5.3. Computational Methods
6. Summary
References and Notes
- Rebek, J., Jr. Angew. Chem. Int. Edit. Engl. 1990, 29, 245. [CrossRef]
- Lehn, J.-M. Angew. Chem. Int. Edit. Engl. 1990, 29, 1304. [CrossRef]
- Schneider, H.-J. Angew. Chem. Int. Edit. Engl. 1991, 30, 1417. [CrossRef]
- Diederich, F. N. Cyclophanes; Royal Society of Chemistry: Cambridge, 1991. [Google Scholar]
- Vögtle, F. Supramolecular Chemistry; Wiley: New York, 1991. [Google Scholar]
- Anelli, P. L.; Ashton, P. R.; Ballardini, R.; Balzani, V.; Delgado, M.; Gandolfi, M. T.; Goodnow, T. T.; Kaifer, A. E.; Philp, D.; Pietraszkiewicz, M.; Prodi, L.; Reddington, M. V.; Slawin, A. M. Z.; Spencer, N.; Stoddart, J. F.; Vicent, C.; Williams, D. J. J. Am. Chem. Soc. 1992, 114, 193.
- Seel, C.; Vögtle, F. Angew. Chem. Int. Edit. Engl. 1992, 31, 528. [CrossRef]
- Davis, A. P. Chem. Soc. Rev. 1993, 22, 243. [CrossRef]
- Lehn, J.-M. Supramolecular Chemistry; VCH: Weinheim, 1995. [Google Scholar]
- Lehn, J.-M.; Atwood, J. L.; Davies, J. E. D.; MacNicol, D. D.; Vögtle, F. (Eds.) Comprehensive Supramolecular Chemistry; Pergamon: Oxford, 1996.
- Li, Y.; Dias, J. R. Chem. Rev. 1997, 97, 283.
- Wallimann, P.; Marti, T.; Fürer, A.; Diederich, F. Chem. Rev. 1997, 97, 1567.
- Antonisse, M. M. G.; Reinhoudt, D. N. Chem. Commun. 1998, 443.
- Diederich, F.; Gomez-Lopez, M. Chem. Soc. Rev. 1999, 28, 263. [CrossRef]
- MacGillivray, L. R.; Atwood, J. L. Angew. Chem. Int. Edit. Engl. 1999, 38, 1018. [CrossRef]
- Davis, A. P.; Wareham, R. S. Angew. Chem. Int. Edit. Engl. 1999, 38, 2978.
- Newkome, G. R.; He, E.; Moorefield, C. N. Chem. Rev. 1999, 99, 1689.
- Rudkevich, D. M.; Rebek, J., Jr. Eur. J. Org. Chem. 1999, 1991.
- Rebek, J., Jr. Chem. Commun. 2000, 637.
- Schneider, H.-J.; Eblinger, F.; Sirish, M. Adv. Supramol. Chem. 2000, 6, 185.
- Philp, D.; Stoddart, J. F. Angew. Chem. Int. Edit. Engl. 1996, 35, 1155.
- Gomez-Lopez, M.; Stoddart, J. F. Bull. Soc. Chim. Belg. 1997, 106, 491.
- Balzani, V.; Gomez-Lopez, M.; Stoddart, J. F. Acc. Chem. Res. 1998, 31, 405. [CrossRef]
- Sauvage, J.-P. Acc. Chem. Res. 1998, 31, 611. [CrossRef]
- Mao, C.; Sun, W.; Shen, Z.; Seeman, N. C. Nature 1999, 397, 144. [PubMed]
- Davis, A. P. Nature 1999, 401, 120. [PubMed]
- Kelly, T. R.; De Silva, H.; Silva, R. A. Nature 1999, 401, 150.
- Koumura, N.; Zijlstra, R. W. J.; van Delden, R. A.; Harada, N.; Feringa, B. L. Nature 1999, 401, 152.
- Raymo, F. M.; Stoddart, J. F. Chem. Rev. 1999, 99, 1643.
- Feringa, B. L.; van Delden, R. A.; Koumura, N.; Geertsema, E. M. Chem. Rev. 2000, 100, 1789.
- Davis, A. P.; Bonar-Law, R. P.; Sanders, J. K. M. Comprehensive Supramolecular Chemistry; Atwood, J. L., Davis, J. E. D., Macnicol, D. D., Vögtle, F., Eds.; Elsevier: Oxford, 1996; Vol. 4, pp. 257–286. [Google Scholar]
- Miyata, M.; Sada, K. Comprehensive Supramolecular Chemistry; Atwood, J. L., Davis, J. E. D., Macnicol, D. D., Vögtle, F., Eds.; Elsevier: Oxford, 1996; Vol. 6, pp. 147–176. [Google Scholar]
- Tamminen, J. PhD Thesis, Department of Chemistry, University of Jyväskylä, Research Report No. 78, Jyväskylä, 2000.
- Gao, H.; Dias, J. R. Org. Prep. Proced. Int. 1999, 31, 145. [CrossRef]
- Kritchevsky, D.; Nair, P. P. The Bile Acids: Chemistry, Physiology, and Metabolism; Nair, P. P., Kritchevsky, D., Eds.; Plenum: New York, 1971; Vol. 1, p. 3. [Google Scholar]
- Enhsen, A.; Kramer, W.; Wess, G. Drug Discovery Today 1998, 3, 409.
- Kramer, W.; Wess, G.; Schubert, G.; Bickel, M.; Girbig, F.; Gutjahr, U.; Kowalewski, S.; Baringhaus, K.-H.; Enhsen, A.; Glombik, H.; Müllner, S.; Neckermann, G.; Schulz, S.; Petzinger, E. J. Biol Chem. 1992, 267, 18598. [PubMed]
- Hofmann, A. F. News Physiol. Sci. 1999, 14, 24. [PubMed]
- Hofmann, A. F. Ital. J. Gastroenterol. 1995, 27, 106. [PubMed]
- Berlati, F.; Ceschel, G.; Clerici, C.; Pellicciari, R.; Roda, A.; Ronchi, C. WO 9400126, 1994.
- Marples, B. A.; Stretton, R. J. WO 9013298, 1990.
- Berlati, F.; Ceschel, G.; Roda, A.; Roda, E.; Ronchi, C. WO 9400155, 1994.
- Swaan, P. W.; Hillgren, K. M.; Szoka, F. C., Jr; Øie, S. Bioconjug. Chem. 1997, 8, 520. [PubMed]
- Sliedregt, L. A. J. M.; Rensen, P. C. N.; Rump, E. T.; van Santbrink, P. J.; Bijsterbosch, M. K.; Valentijn, A. R. P. M.; van der Marel, G. A.; van Boom, J. H.; van Berkel, T. J. C.; Biessen, E. A. L. J. Med Chem. 1999, 42, 609. [PubMed]
- Kramer, W.; Glombik, H. DE 19824123A1, 1999.
- Geall, A. J.; Al-Hadithi, D.; Blagbrough, I. S. Chem. Commun. 1998, 2035.
- Ruff, M. R.; Hill, J. M.; Kwart, L. D.; Pert, C. B. US 5446026, 1995.
- Li, C.; Peters, A. S.; Meredith, E. L.; Allman, G. W.; Savage, P. B. J. Am. Chem. Soc. 1998, 120, 2961.
- Li, C.; Budge, L. P.; Driscoll, C. D.; Willardson, B. M.; Allman, G. W.; Savage, P. S. J. Am. Chem. Soc. 1999, 121, 931.
- Campazzi, E.; Cattabriga, M.; Marvelli, L.; Marchi, A.; Rossi, R.; Pieragnoli, M. R.; Fogagnolo, M. Inorg. Chim. Acta 1999, 286, 46.
- Bell, D. A.; Anslyn, E. V. Comprehensive Supramolecular Chemistry; Atwood, J. L., Davis, J. E. D., Macnicol, D. D., Vögtle, F., Eds.; Elsevier: Oxford, 1996; Vol. 2, pp. 439–475. [Google Scholar]
- Inanaga, J.; Hirata, K.; Saeki, H.; Katsuki, T.; Yamaguchi, M. Bull. Chem. Soc. Jpn. 1979, 52, 1989. [CrossRef]
- Lappalainen, K.; Kolehmainen, E.; Šaman, D. Spectrochim. Acta, Part A 1995, 51, 1543. [CrossRef]
- Lappalainen, K.; Kolehmainen, E.; Kotoneva, J. Magn. Res. Chem. 1996, 34, 316. [CrossRef]
- Lappalainen, K.; Kolehmainen, E. Liebigs Annalen-Recueil 1997, 1965.
- Bonar-Law, R. P.; Sanders, J. K. M. Tetrahedron Lett. 1992, 33, 2071.
- Bonar-Law, R. P.; Sanders, J. K. M. Tetrahedron Lett. 1993, 34, 1677.
- Li, Y.; Dias, J. R. Synthesis 1997, 425.
- Gao, H.; Dias, J. R. Synth. Commun. 1997, 27, 757.
- Gao, H.; Dias, J. R. New J. Chem. 1998, 579.
- Gao, H.; Dias, J. R. Eur. J. Org. Chem. 1998, 719.
- Gao, H.; Dias, J. R. Croatica Chemica Acta 1998, 71, 827.
- Bonar-Law, R. P.; Sanders, J. K. M. J. Chem. Soc., Chem. Commun. 1991, 574.
- Bonar-Law, R. P.; Mackay, L. G.; Sanders, J. K. M. J. Chem. Soc., Chem. Commun. 1993, 456.
- Mackay, L. G.; Bonar-Law, R. P.; Sanders, J. K. M. J. Chem. Soc., Perkin Trans. 1 1993, 1377.
- Bonar-Law, R. P.; Mackay, L. G.; Walter, C. J.; Marvaud, V.; Sanders, J. K. M. Pure Appl. Chem. 1994, 66, 803.
- Bonar-Law, R. P.; Sanders, J. K. M. J. Am. Chem. Soc. 1995, 117, 259.
- Bonar-Law, R. P.; Sanders, J. K. M. J. Chem. Soc., Perkin Trans. 1 1995, 3085.
- Bonar-Law, R. P. J. Am. Chem. Soc. 1995, 117, 12397.
- Brady, P. A.; Bonar-Law, R. P.; Rowan, S. J.; Suckling, C. J.; Sanders, J. K. M. Chem. Commun. 1996, 319.
- Brady, P. A.; Sanders, J. K. M. J. Chem. Soc., Perkin Trans. 1. 1997, 3237.
- Brady, P. A.; Sanders, J. K. M. New J. Chem. 1998, 411.
- Bonar-Law, R. P.; Davis, A. P. J. Chem. Soc., Chem. Commun. 1989, 1050.
- Bonar-Law, R. P.; Davis, A. P.; Murray, B. A. Angew. Chem. Int. Edit. Engl. 1990, 29, 1407. [CrossRef]
- Bhattarai, K. M.; Bonar-Law, R. P.; Davis, A. P.; Murray, B. A. J. Chem. Soc., Chem. Commun. 1992, 752.
- Bonar-Law, R. P.; Davis, A. P. Tetrahedron 1993, 49, 9829. Bonar-Law, R. P.; Davis, A. P. Tetrahedron 1993, 49, 9845. Bonar-Law, R. P.; Davis, A. P.; Dorgan, B. J. Tetrahedron 1993, 49, 9855.
- Davis, A. P.; Walsh, J. J. Chem. Commun. 1996, 449.
- Davis, A. P.; Menzer, S.; Walsh, J. J.; Williams, D. J. Chem. Commun. 1996, 453.
- Bhattarai, K. M.; Davis, A. P.; Perry, J. J.; Walter, C. J. J. Org. Chem. 1997, 62, 8463.
- Albert, D.; Feigel, M. Tetrahedron Lett. 1994, 35, 565.
- Albert, D.; Feigel, M. Helv. Chim. Acta 1997, 80, 2168. [CrossRef]
- Albert, D.; Feigel, M.; Benet-Buchholz, J.; Boese, R. Angew. Chem. Int. Edit. Engl. 1998, 37, 2727. [CrossRef]
- Pandey, P. P.; Singh, R. B. Tetrahedron Lett. 1997, 38, 5045.
- Kolehmainen, E.; Tamminen, J.; Lappalainen, K.; Torkkel, T.; Seppälä, R. Synthesis 1996, 1082.
- Tamminen, J.; Kolehmainen, E.; Haapala, M.; Linnanto, J. Synthesis 2000, 1464.
- Haapala, M.; Kolehmainen, E.; Tamminen, J.; Kauppinen, R.; Linnanto, J.; Virtanen, E.; Suontamo, R.; Vainiotalo, P. Mater. Sci. Eng., C. in press.
- Maitra, U.; Balasubramanian, S. J. Chem. Soc., Perkin Trans. 1 1995, 83.
- Maitra, U. Curr. Sci. 1996, 71, 617.
- Maitra, U.; Bag, B. G. J. Org. Chem. 1994, 59, 6114.
- Maitra, U.; D'Souza, L. J.; Kumar, P. V. Supramol. Chem. 1998, 10, 97.
- Kohmoto, S.; Fukui, D.; Nagashima, T.; Kishikawa, K.; Yamamoto, M.; Yamada, K. Chem. Commun. 1996, 1869.
- Irie, S.; Yamamoto, M.; Kishikawa, K.; Kohmoto, S.; Yamada, K. Synthesis 1996, 1135.
- Kikuchi, J.-I.; Murakami, Y. J. Inclusion Phenom. Mol. Recognit. Chem. 1998, 32, 209. [CrossRef]
- Davis, A. P.; Gilmer, J. F.; Perry, J. J. Angew. Chem. Int. Edit. Engl. 1996, 35, 1312. [CrossRef]
- Kohmoto, S.; Sakayori, K.; Kishikawa, K.; Yamamoto, M. J. Chem. Soc., Perkin Trans. 2 1999, 833.
- McKenna, J.; McKenna, J. M.; Thornthwaite, D. W. Chem. Soc., Chem. Commun. 1977, 809.
- Tamminen, J.; Lappalainen, K.; Laihia, K.; Mänttäri, P.; Salo, H.; Kolehmainen, E. Magn. Res. Chem. 1999, 37, 163. [CrossRef]
- Kolehmainen, E.; Tamminen, J.; Kauppinen, R.; Linnanto, J. J. Inclusion Phenom. Macrocyclic Chem. 1999, 35, 75. [CrossRef]
- Tamminen, J.; Kolehmainen, E.; Linnanto, J.; Salo, H.; Mänttäri, P. Magn. Res. Chem. 2000, 38, 877. [CrossRef]
- Maitra, U.; D'Souza, L. J. J. Chem. Soc., Chem. Commun. 1994, 2793.
- D'Souza, L. J.; Maitra, U. J. Org. Chem. 1996, 61, 9494.
- Maitra, U.; Rao, P.; Kumar P, V.; Balasubramanian, R.; Mathew, L. Tetrahedron Lett. 1998, 39, 3255.
- Rao, P.; Maitra, U. Supramol. Chem. 1998, 9, 325.
- Davis, A. P.; Perry, J. J.; Williams, R. P. J. Am. Chem. Soc. 1997, 119, 1793.
- Davis, A. P.; Perry, J. J.; Wareham, R. S. Tetrahedron Lett. 1998, 39, 4569.
- Broderick, S.; Davis, A. P.; Williams, R. P. Tetrahedron Lett. 1998, 39, 6083.
- Davis, A. P.; Dresen, S.; Lawless, L. J. Tetrahedron Lett. 1997, 38, 4305.
- Davis, A. P.; Pérez-Payán, M. N. Synlett 1999, 991.
- Li, C.; Ur-Rehman, A.; Dalley, N. K.; Savage, P. B. Tetrahedron Lett. 1999, 40, 1861.
- Vandenburg, Y. R.; Smith, B. D.; Pérez-Payán, M. N.; Davis, A. P. J. Am. Chem. Soc. 2000, 122, 3252.
- Davis, A. P.; Lawless, L. J. Chem. Commun. 1999, 9.
- Balasubramanian, R.; Rao, P.; Maitra, U. Chem. Commun. 1999, 2353.
- Maitra, U.; Kumar, P. V.; Chandra, N.; D’Souza, L. J.; Prasanna, M. D.; Raju, A. R. Chem. Commun. 1999, 595.
- Tamminen, J.; Kolehmainen, E.; Haapala, M.; Salo, H.; Linnanto, J. ARKIVOC 2000, 1, 90.
- Tamminen, J.; Kolehmainen, E.; Linnanto, J.; Vainiotalo, P.; Vuorikoski, S.; Kauppinen, R. J. Inclusion Phenom. Macrocyclic Chem. 2000, 37, 121. [CrossRef]
- Zuluaga, F.; Valderruten, N. E.; Wagener, K. B. Polym. Bull. 1999, 42, 41.
- Zhang, Y. H.; Akram, M.; Liu, H. Y.; Zhu, X. X. Macromol. Chem. Phys. 1998, 199, 1399.
- Goto, J.; Murao, N.; Oohashi, J.; Ikegawa, S. Steroids 1998, 63, 180.
- Janout, V.; Lanier, M.; Regen, S. L. J. Am. Chem. Soc. 1996, 118, 1573.
- Janout, V.; Lanier, M.; Regen, S. L. J. Am. Chem. Soc. 1997, 119, 640.
- Miyata, M.; Shibakami, M.; Goonewardena, W.; Takemoto, K. Chem. Lett. 1987, 605.
- Miyata, M.; Shibakami, M.; Chirachanchai, S.; Takemoto, K.; Kasai, N.; Miki, K. Nature 1990, 343, 446.
- Sada, K.; Shiomi, N.; Miyata, M. J. Am. Chem. Soc. 1998, 120, 10543.
- Hishikawa, Y.; Aoki, Y.; Sada, K.; Miyata, M. Chem. Lett. 1998, 1289.
- Hishikawa, Y.; Watanabe, R.; Sada, K.; Miyata, M. Chirality 1998, 10, 600.
- Miyake, Y.; Hirose, J.; Hasegawa, Y.; Sada, K.; Miyata, M. Chem. Commun. 1998, 111.
- Sugahara, M.; Sada, K.; Miyata, M. Chem. Commun. 1999, 293.
- Gdaniec, M.; Polonski, T. J. Am. Chem. Soc. 1998, 120, 7353.
- Gdaniec, M.; Milewska, M. J.; Polonski, T. Angew. Chem. Int. Edit. Engl. 1999, 38, 392. [CrossRef]
- Boyce, R.; Li, G.; Nestler, H. P.; Suenaga, T.; Still, W. C. J. Am. Chem. Soc. 1994, 116, 7955.
- Cheng, Y.; Suenaga, T.; Still, W. C. J. Am. Chem. Soc. 1996, 118, 1813.
- Wess, G.; Bock, K.; Kleine, H.; Kurz, M.; Guba, W.; Hemmerle, H.; Lopez-Calle, E.; Baringhaus, K.-H.; Glombik, H.; Enhsen, A.; Kramer, W. Angew. Chem. Int. Edit. Engl. 1996, 35, 2222. [CrossRef]
- Barry, J. F.; Davis, A. P.; Nieves Pérez-Payan, M.; Elsegood, M. R. J.; Jackson, R. F. W.; Gennari, C.; Piarulli, U.; Gude, M. Tetrahedron Lett. 1996, 40, 2849.
- De Muynck, H.; Madder, A.; Farcy, N.; De Clercq, P. J.; Nieves Pérez-Payán, M.; Öhberg, L. M.; Davis, A. P. Angew. Chem. Int. Edit. Engl. 2000, 39, 145. [CrossRef]
- Derome, A. E.; Williamson, M. P. J. Magn. Reson. 1990, 88, 177.
- Kirk, D. N.; Toms, H. C.; Douglas, C.; White, K. A.; Smith, K. E.; Latif, S.; Hubbard, R. W. P. J. Chem. Soc., Perkin Trans. 2 1990, 1567.
- Iida, T.; Chang, F. C.; Mushiake, K.; Goto, J.; Nambara, T. Magn. Res. Chem. 1993, 31, 645. [CrossRef]
- Liu, M.; Farrant, R. D.; Lindon, J. C.; Nicholson, J. K. Magn. Res. Chem. 1995, 33, 212. [CrossRef]
- Momose, T.; Iida, T.; Mushiake, K.; Goto, J.; Nambara, T. Magn. Res. Chem. 1996, 34, 681. [CrossRef]
- Reynolds, W. F.; McLean, S.; Tay, L.-L.; Yu, M.; Enriquez, R. G.; Estwick, D. M.; Pascoe, K. O. Magn. Res. Chem. 1997, 35, 455. [CrossRef]
- Yim, C. T.; Zhu, X. X.; Brown, G. R. J. Phys. Chem. B 1999, 103, 597. [CrossRef]
- Bortolini, O.; Carpi, A.; Fantin, G.; Guerrini, A.; Medici, A. Ann. Chim. 1999, 89, 319.
- Iida, T.; Komatsubara, I.; Yoda, S.; Goto, J.; Nambara, T.; Chang, F. C. Steroids 1990, 55, 530.
- Blunt, J. W.; Stothers, J. B. Org. Magn. Reson. 1977, 9, 439.
- Iida, T.; Tamura, T.; Matsumoto, T.; Chang, F. C. Org. Magn. Reson. 1983, 21, 305.
- Iida, T.; Chang, F. C.; Goto, J.; Nambara, T. Chem. Phys. Lipids 1987, 45, 1. [CrossRef]
- Iida, T.; Goto, J.; Nambara, T. Magn. Res. Chem. 1993, 31, 421. [CrossRef]
- Blossey, E. C.; Ford, W. T.; Periyasamy, M. Magn. Res. Chem. 1991, 29, 190. [CrossRef]
- Dias, J. R.; Gao, H.; Kolehmainen, E. Spectrochim. Acta, Part A 2000, 56, 53.
- Kolehmainen, E.; Kaartinen, M.; Kauppinen, R.; Kotoneva, J.; Lappalainen, K.; Lewis, P. T.; Seppälä, R.; Sundelin, J.; Vatanen, V. Magn. Res. Chem. 1994, 32, 441. [CrossRef]
- Smith, L. L.; Herz, J. E.; Ezell, E. L. Steroids 1993, 58, 260.
- Lawson, A. M.; Setchell, K. D. R. The Bile Acids: Chemistry, Physiology, and Metabolism; Setchell, K. D. R., Kritchevsky, D., Nair, P. P., Eds.; Plenum: New York, London, 1988; Vol. 4, pp. 167–267. [Google Scholar]
- Dias, J. R.; Nassim, B. Org. Mass Spectrom. 1978, 13, 402.
- Dias, J. R.; Nassim, B. J. Org. Chem. 1980, 45, 337.
- SPARTAN, Version 5.0, Wavefunction Inc., Irvine, CA (1993-1997).
- Frisch, M. J.; Trucks, G. W.; Schlegel, H. B.; et al. GAUSSIAN 94; Revision B.1; Gaussian Inc.: Pittsburgh PA, (1995).
- Frisch, M. J.; Trucks, G. W.; Schlegel, H. B.; et al. GAUSSIAN 98; Revision A.6; Gaussian Inc.: Pittsburgh PA, (1998).
- Lampert, H.; Mikenda, W.; Karpfen, A.; Kählig, H. J. Phys. Chem. A 1997, 101, 9610.
- Cammi, R.; Mennucci, B.; Tomasi, J. J. Chem. Phys. 1999, 110, 7627.
- Smith, W. B. Magn. Res. Chem. 1999, 37, 103. [CrossRef]
- Samples Availability: Not applicable.
© 2001 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Tamminen, J.; Kolehmainen, E. Bile Acids as Building Blocks of Supramolecular Hosts. Molecules 2001, 6, 21-46. https://doi.org/10.3390/60100021
Tamminen J, Kolehmainen E. Bile Acids as Building Blocks of Supramolecular Hosts. Molecules. 2001; 6(1):21-46. https://doi.org/10.3390/60100021
Chicago/Turabian StyleTamminen, Jari, and Erkki Kolehmainen. 2001. "Bile Acids as Building Blocks of Supramolecular Hosts" Molecules 6, no. 1: 21-46. https://doi.org/10.3390/60100021