Homologous NF-YC2 Subunit from Arabidopsis and Tobacco Is Activated by Photooxidative Stress and Induces Flowering
Abstract
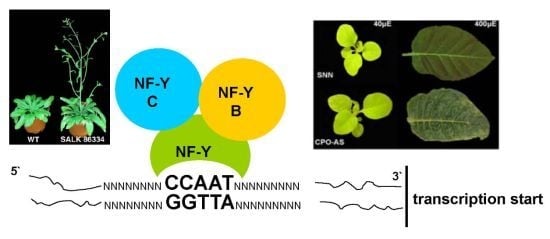
:1. Introduction
2. Results
2.1. Identification of Early Inducible Genes in Response to Photooxidative Stress Triggered by Accumulation of Coproporphyrin in Tobacco
2.2. Transcript Analysis of 36 Arabidopsis Genes Encoding NF-Y Subunits
2.3. An Arabidopsis nf-yc2 Mutant and NF-YC2 Overexpressor Plants upon Oxidative Stress
2.4. Characterization of Other nf-yc Mutants Related to nf-yc2
2.5. Modified Flowering Time of NF-YC1 and NF-YC2 Overexpressor Lines
2.6. Modified Expression of Genes Involved in Flowering Induction in 35SCaMV:NF-YC2 and 35SCaMV:NF-YC1 Overexpressor Lines
3. Discussion
4. Material and Methods
4.1. Growth of Plants
4.2. RNA—Quantification
4.3. Plant Mutants
4.4. Chlorophyll Fluorescence Analyses
4.5. Analysis of Porphyrins
4.6. Suppression Subtractive Hybridization
4.7. Statistical Analysis
5. Conclusion
Acknowledgements
References
- Mantovani, R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res 1998, 26, 1135–1143. [Google Scholar]
- Rieping, M.; Schoffl, F. Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Mol. Gen. Genet 1992, 231, 226–232. [Google Scholar]
- Mantovani, R. The molecular biology of the CCAAT-binding factor NF-Y. Gene 1999, 239, 15–27. [Google Scholar]
- Maity, S.N.; de Crombrugghe, B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci 1998, 23, 174–178. [Google Scholar]
- Matuoka, K.; Chen, K.Y. Nuclear factor Y (NF-Y) and cellular senescence. Exp. Cell Res 1999, 253, 365–371. [Google Scholar]
- Li, X.Y.; Mantovani, R.; van Huijsduijnen, R.H.; Andre, I.; Benoist, C.; Mathis, D. Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Res 1992, 20, 1087–1091. [Google Scholar]
- Liberati, C.; di Silvio, A.; Ottolenghi, S.; Mantovani, R. NF-Y binding to twin CCAAT boxes: Role of Q-rich domains and histone fold helices. J. Mol. Biol 1999, 285, 1441–1455. [Google Scholar]
- Masiero, S.; Imbriano, C.; Ravasio, F.; Favaro, R.; Pelucchi, N.; Gorla, M.S.; Mantovani, R.; Colombo, L.; Kater, M.M. Ternary complex formation between MADS-box transcription factors and the histone fold protein NF-YB. J. Biol. Chem 2002, 277, 26429–26435. [Google Scholar]
- Sinha, S.; Maity, S.N.; Lu, J.; de Crombrugghe, B. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc. Natl. Acad. Sci. USA 1995, 92, 1624–1628. [Google Scholar]
- Gusmaroli, G.; Tonelli, C.; Mantovani, R. Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 2002, 283, 41–48. [Google Scholar]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.T.; Tayrose, G.; Holt, B.F., III. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 2009, 149, 625–641. [Google Scholar]
- Gusmaroli, G.; Tonelli, C.; Mantovani, R. Regulation of the CCAAT-Binding NF-Y subunits in Arabidopsis thaliana. Gene 2001, 264, 173–185. [Google Scholar]
- Hackenberg, D.; Wu, Y.; Voigt, A.; Adams, R.; Schramm, P.; Grimm, B. Studies on differential nuclear translocation mechanism and assembly of the three subunits of the arabidopsis thaliana transcription factor NF-Y. Mol. Plant 2011. [Google Scholar] [CrossRef]
- Lotan, T.; Ohto, M.; Yee, K.M.; West, M.A.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar]
- Kwong, R.W.; Bui, A.Q.; Lee, H.; Kwong, L.W.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 2003, 15, 5–18. [Google Scholar]
- Folta, K.M.; Kaufman, L.S. Regions of the pea Lhcb1*4 promoter necessary for blue-light regulation in transgenic Arabidopsis. Plant Physiol 1999, 120, 747–756. [Google Scholar]
- Kumimoto, R.W.; Adam, L.; Hymus, G.J.; Repetti, P.P.; Reuber, T.L.; Marion, C.M.; Hempel, F.D.; Ratcliffe, O.J. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar]
- Kumimoto, R.W.; Zhang, Y.; Siefers, N.; Holt, B.F. NF–YC3, NF–YC4 and NF–YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J 2010, 63, 379–391. [Google Scholar]
- Tiwari, S.B.; Shen, Y.; Chang, H.C.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; Belachew, A.; Repetti, P.P.; Reuber, T.L.; Ratcliffe, O.J. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 2010, 187, 57–66. [Google Scholar]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar]
- Liu, J.X.; Howell, S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar]
- Kruse, E.; Mock, H.P.; Grimm, B. Reduction of coproporphyrinogen oxidase level by antisense RNA synthesis leads to deregulated gene expression of plastid proteins and affects the oxidative defense system. EMBO J 1995, 14, 3712–3720. [Google Scholar]
- Keetman, U.; Mock, H.P.; Grimm, B. Kinetics of antioxidative defence responses to photosensitisation in porphyrin-accumulating tobacco plants. Plant Physiol. Biochem 2002, 40, 697–707. [Google Scholar]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the nonspecialist. Nucleic Acids Res 2008, 36, W465–469. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar]
- Witkowski, D.A.; Halling, B.P. Inhibition of plant protoporphyrinogen oxidase by the herbicide acifluorfen-methyl. Plant Physiol 1989, 90, 1239–1242. [Google Scholar]
- Shigeoka, S.; Ishikawa, T.; Tamoi, M.; Miyagawa, Y.; Takeda, T.; Yabuta, Y.; Yoshimura, K. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot 2002, 53, 1305–1319. [Google Scholar]
- Koussevitzky, S.; Suzuki, N.; Huntington, S.; Armijo, L.; Sha, W.; Cortes, D.; Shulaev, V.; Mittler, R. Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J. Biol. Chem 2008, 283, 34197–34203. [Google Scholar]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar]
- Jin, H.; Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol 1999, 41, 577–585. [Google Scholar]
- Turck, F.; Fornara, F.; Coupland, G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol 2008, 59, 573–594. [Google Scholar]
- Ben-Naim, O.; Eshed, R.; Parnis, A.; Teper-Bamnolker, P.; Shalit, A.; Coupland, G.; Samach, A.; Lifschitz, E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J 2006, 46, 462–476. [Google Scholar]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; le Gourrierec, J.; Samach, A.; Coupland, G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar]
- Turnbull, C. Long-distance regulation of flowering time. J. Exp. Bot 2011, 62, 4399–4413. [Google Scholar]
- Jang, S.; Torti, S.; Coupland, G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J 2009, 60, 614–625. [Google Scholar]
- Rieu, I.; Ruiz-Rivero, O.; Fernandez-Garcia, N.; Griffiths, J.; Powers, S.J.; Gong, F.; Linhartova, T.; Eriksson, S.; Nilsson, O.; Thomas, S.G.; et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 2008, 53, 488–504. [Google Scholar]
- Hisamatsu, T.; King, R.W. The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J. Exp. Bot 2008, 59, 3821–3829. [Google Scholar]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett 2003, 339, 62–66. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998, 16, 735–743. [Google Scholar]
- Russel, S. Molecular Cloning: A Laboratory Manual, 3rd ed; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Lokstein, H.; Tian, L.; Polle, J.E.; DellaPenna, D. Xanthophyll biosynthetic mutants of Arabidopsis thaliana: Altered nonphotochemical quenching of chlorophyll fluorescence is due to changes in Photosystem II antenna size and stability. Biochim. Biophys. Acta 2002, 1553, 309–319. [Google Scholar]
- Rohacek, K. Chlorophyll fluorescence parameters: The definitions, photosynthetic meaning, and mutual relationships. Photosynthetica 2002, 40, 13–29. [Google Scholar]






| Stress condition | AtNF-Y with increased expression | AtNF-Y with lower expression | ||
|---|---|---|---|---|
| AtNF-Y | relative expression | AtNF-Y | relative expression | |
| 10 mM H2O2 | ATNF-YA1 | 5.0 (1.6–15.8) | ATNF-YB7 | 0.3 (0.2–0.7) |
| ATNF-YC2 | 3.1 (1.9–5.1) | ATNF-YC12 | 0.5 (0.4–0.7) | |
| 20 μM Norflurazon | ATNF-YA1 | 2.3 (1.3–3.9) | ATNF-YB1 | 0.4 (0.2–0.7) |
| ATNF-YA10 | 3.1 (2.0–4.9) | ATNF-YB6 | 0.4 (0.2–0.6) | |
| ATNF-YC2 | 2.9 (1.7–5.1) | ATNF-YC12 | 0.5 (0.3–0.8) | |
| 50 μM Acifluorfen | ATNF-YC2 | 5.8 (4.7–7.1) | ATNF-YA2 | 0.5 (0.2–1.3) |
| ATNF-YB6 | 0.3 (0.3–0.4) | |||
| ATNF-YB7 | 0.2 (0.2–0.2) | |||
| ATNF-YC12 | 0.4 (0.2–0.6) | |||
| High light 500 μE | ATNF-YA1 | 2.1 (1.1–4.0) | ATNF-YC1 | 0.4 (0.2–0.6) |
| ATNF-YC2 | 4.0 (1.9–8.3) | ATNF-YC4 | 0.1 (0.1–0.3) | |
| ATNF-YC8 | 0.4 (0.1–1.1) | |||
| Low light 20 μE | ATNF-YC4 | 2.1 (1.2–3.5) | ATNF-YC2 | 0.5 (0.4–0.7) |
| ATNF-YC13 | 0.4 (0.3–0.5) | |||
| Heat stress 38 °C | ATNF-YA1 | 4.2 (2.6–6.8) | ATNF-YB7 | 0.3 (0.2–0.5) |
| ATNF-YA5 | 2.5 (1.7–3.7) | ATNF-YC13 | 0.1 (0.0–0.1) | |
| ATNF-YA7 | 2.3 (1.2–4.6) | |||
| ATNF-YB13 | 2.5 (1.4–4.4) | |||
| ATNF-YB6 | 3.6 (1.9–6.6) | |||
| ATNF-YB8 | 4.1 (2.7–6.3) | |||
| ATNF-YC10 | 2.1 (1.3–3.3) | |||
| ATNF-YC2 | 4.8 (2.5–9.0) | |||
| ATNF-YC4 | 9.4 (6.6–134) | |||
| Cold stress 4 °C | ATNF-YB2 | 0.4 (0.3–0.5) | ||
| ATNF-YB3 | 0.2 (0.2–0.4) | |||
| ATNF-YB8 | 0.4 (0.2–0.7) | |||
| ATNF-YC12 | 0.4 (0.2–0.6) | |||
| ATNF-YC4 | 0.4 (0.2–0.6) | |||
| Drought stress | ATNF-YA1 | 21.7 (7.3–64.6) | ATNF-YC1 | 0.4 (0.2–0.8) |
| ATNF-YA10 | 48.0 (6.7–345.6) | |||
| ATNF-YA3 | 5.9 (3.1–11.4) | |||
| ATNF-YA4 | 5.4 (3.6–8.1) | |||
| ATNF-YA5 | 9.7 (4.1–23.1) | |||
| ATNF-YA7 | 7.9 (5.4–11.5) | |||
| ATNF-YB1 | 9.0 (5.0–16.0) | |||
| ATNF-YB10 | 22.6 (8.9–57.7) | |||
| ATNF-YB6 | 57.4 (30.7–107.3) | |||
| ATNF-YB7 | 5.2 (2.8–9.5) | |||
| ATNF-YC10 | 15.2 (9.0–25.8) | |||
| ATNF-YC2 | 6.2 (4.0–9.5) | |||
| ATNF-YC3 | 7.6 (4.3–13.4) | |||
| ATNF-YC4 | 5.7 (3.3–10.2) | |||
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hackenberg, D.; Keetman, U.; Grimm, B. Homologous NF-YC2 Subunit from Arabidopsis and Tobacco Is Activated by Photooxidative Stress and Induces Flowering. Int. J. Mol. Sci. 2012, 13, 3458-3477. https://doi.org/10.3390/ijms13033458
Hackenberg D, Keetman U, Grimm B. Homologous NF-YC2 Subunit from Arabidopsis and Tobacco Is Activated by Photooxidative Stress and Induces Flowering. International Journal of Molecular Sciences. 2012; 13(3):3458-3477. https://doi.org/10.3390/ijms13033458
Chicago/Turabian StyleHackenberg, Dieter, Ulrich Keetman, and Bernhard Grimm. 2012. "Homologous NF-YC2 Subunit from Arabidopsis and Tobacco Is Activated by Photooxidative Stress and Induces Flowering" International Journal of Molecular Sciences 13, no. 3: 3458-3477. https://doi.org/10.3390/ijms13033458