Expression and Genetic Analysis of MicroRNAs Involved in Multiple Sclerosis
Abstract
:1. Introduction
2. Results and Discussion
2.1. miR-223 and miR-23a Expression Levels Are Altered in MS Patients versus Controls
2.2. Genetic Analysis of miR-223, miR-23a and miR-15b in MS Patients
3. Experimental Section
3.1. Subjects
3.2. DNA Isolation
3.3. Serum Sampling
3.4. miR-223, miR-23a and miR-15b Extraction from PBMCs and Serum and Quantitative Analysis by Real-Time PCR
3.5. Genotyping
3.6. Statistical Analysis
4. Conclusions and Limitations
Acknowledgments
Conflict of Interest
References
- Miller, A.E. Multiple sclerosis: Where will be in 2020? Mt. Sinai J. Med 2011, 78, 268–279. [Google Scholar]
- Stys, P.K.; Zamponi, G.W.; van Minnen, J.; Geurts, J.J.G. Will the real multiple sclerosis stand up? Nat. Rev. Immunol 2012, 13, 507–514. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Fenoglio, C.; Ridolfi, E.; Galimberti, D.; Scarpini, E. MicroRNAs: Active players in the pathogenesis of Multiple Sclerosis. Int. J. Mol. Sci 2012, 13, 13227–13239. [Google Scholar]
- Du, C.; Liu, C.; Kang, J.; Zhao, G.; Ye, Z.; Huang, S.; Li, Z.; Wu, Z.; Pei, G. MicroRNA miR-326 regulates Th-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol 2009, 10, 1252–1259. [Google Scholar]
- Xiao, C.; Srinivasan, L.; Calado, D.P.; Patterson, H.C.; Zhang, B.; Wang, J.; Henderson, J.M. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol 2008, 9, 405–414. [Google Scholar]
- Guerau-de-Arellano, M.; Smith, K.M.; Godlewsky, J.; Liu, Y.; Winger, R.; Lawler, S.E.; Whitacre, C.C.; Racke, M.K.; Lovett-Racke, A.E. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain 2011, 134, 3578–3589. [Google Scholar]
- Otaegui, D.; Baranzini, S.E.; Armananzas, R.; Calvo, B.; Munoz-Culla, M.; Khankhanian, P.; Inza, I.; Lozano, J.A.; Castillo-Trivino, T.; Asensio, A.; et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS One 2009, 4, e6309. [Google Scholar]
- Martinelli-Boneschi, F.; Fenoglio, C.; Brambilla, P.; Sorosina, M.; Giacalone, G.; Esposito, F.; Serpente, M.; Cantoni, C.; Ridolfi, E.; Rodegher, M.; et al. MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci. Lett 2012, 508, 4–8. [Google Scholar]
- Cox, M.B.; Cairns, M.J.; Gandhi, K.S.; Carroll, A.P.; Moscovis, S.; Stewart, G.J.; Broadley, S.; Scott, R.J.; Booth, D.R.; Lechner-Scott, J.; et al. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS One 2010, 5, e12132. [Google Scholar]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 2009, 132, 3342–3352. [Google Scholar]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar]
- Haghikia, A.; Haghikia, A.; Hellwig, K.; Baraniskin, A.; Holzmann, A.; Décard, B.F.; Thum, T.; Gold, R. Regulated microRNAs in the CSF of patients with multiple sclerosis: A case-control study. Neurology 2012, 79, 2166–2170. [Google Scholar]
- Ajit, S.K. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors 2012, 12, 3359–3369. [Google Scholar]
- Fenoglio, C.; Ridolfi, E.; Cantoni, C.; De Riz, M.; Pietroboni, A.; Serpente, M.; Villa, C.; Bonsi, R.; Bresolin, N.; Piccio, L.; et al. Decreased circulating miRNAs in patients with multiple sclerosis. Mult. Scler. J. 2011, 18(S4), 255–277. [Google Scholar]
- Zampetaki, A.; Willeit, P.; Drozdov, I.; Kiechl, S.; Mayr, M. Profiling of circulating microRNAs: From single biomarkers to re-wired networks. Cardiovasc. Res. 2011, 93. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Lange, J.; Borries, A.; Schroers, H.; Scheffler, H.P.; Lenhof, H.P.; Ruprecht, K.; Meese, E. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing remitting disease from healthy controls. PLoS One 2009, 4, e7440. [Google Scholar]
- De Santis, G.; Ferracin, M.; Biondani, A.; Caniatti, L.; Tola, R.; Castellazzi, M. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J. Neuroimmunol 2010, 226, 165–171. [Google Scholar]
- Redis, R.S.; Calin, S.; Yang, Y.; You, M.J.; Calin, G.A. Cell-to-cell miRNA transfer: From body homeostasis to therapy. Pharmacol. Ther. 2012, 136. [Google Scholar] [CrossRef]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Lui, Y.; Huang, J.D.; Song, E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [Green Version]
- Li, T.; Morgan, M.J.; Choksi, S.; Zhang, Y.; Kim, Y.S.; Liu, Z.G. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat. Immunol 2010, 11, 799–805. [Google Scholar]
- Clemente, D.; Ortega, M.C.; Arenzana, F.J.; de Castro, F. FGF-2 and Anosmin-1 are selectively expressed in different types of multiple sclerosis lesions. J. Neurosci 2011, 31, 14899–14909. [Google Scholar]
- Sarchielli, P.; Di Filippo, M.; Ercolani, M.V.; Chiasserini, D.; Mattioni, A. Fibroblast growth factor-2 levels are elevated in the cerebrospinal fluid of multiple sclerosis patients. Neurosci. Lett 2008, 435, 223–228. [Google Scholar]
- Lin, S.T.; Fu, Y.H. MiR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis. Model Mech 2009, 2, 178–188. [Google Scholar]
- Vogt, L.; Schmitz, N.; Kurrer, M.O.; Bauer, M.; Hinton, H.I.; Behnke, S.; Gatto, D.; Sebbel, P.; Beerli, R.R.; Sonderegger, I.; et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J. Clin. Invest 2006, 116, 2817–2824. [Google Scholar]
- Polman, C.H.; Reingold, S.C.; Edan, G.; Filippi, M.; Harthung, H.P.; Kappos, L.; Lublin, F.D.; Metz, L.M.; McFarland, H.F.; O’Connor, P.W.; et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann. Neurol 2005, 58, 840–846. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar]
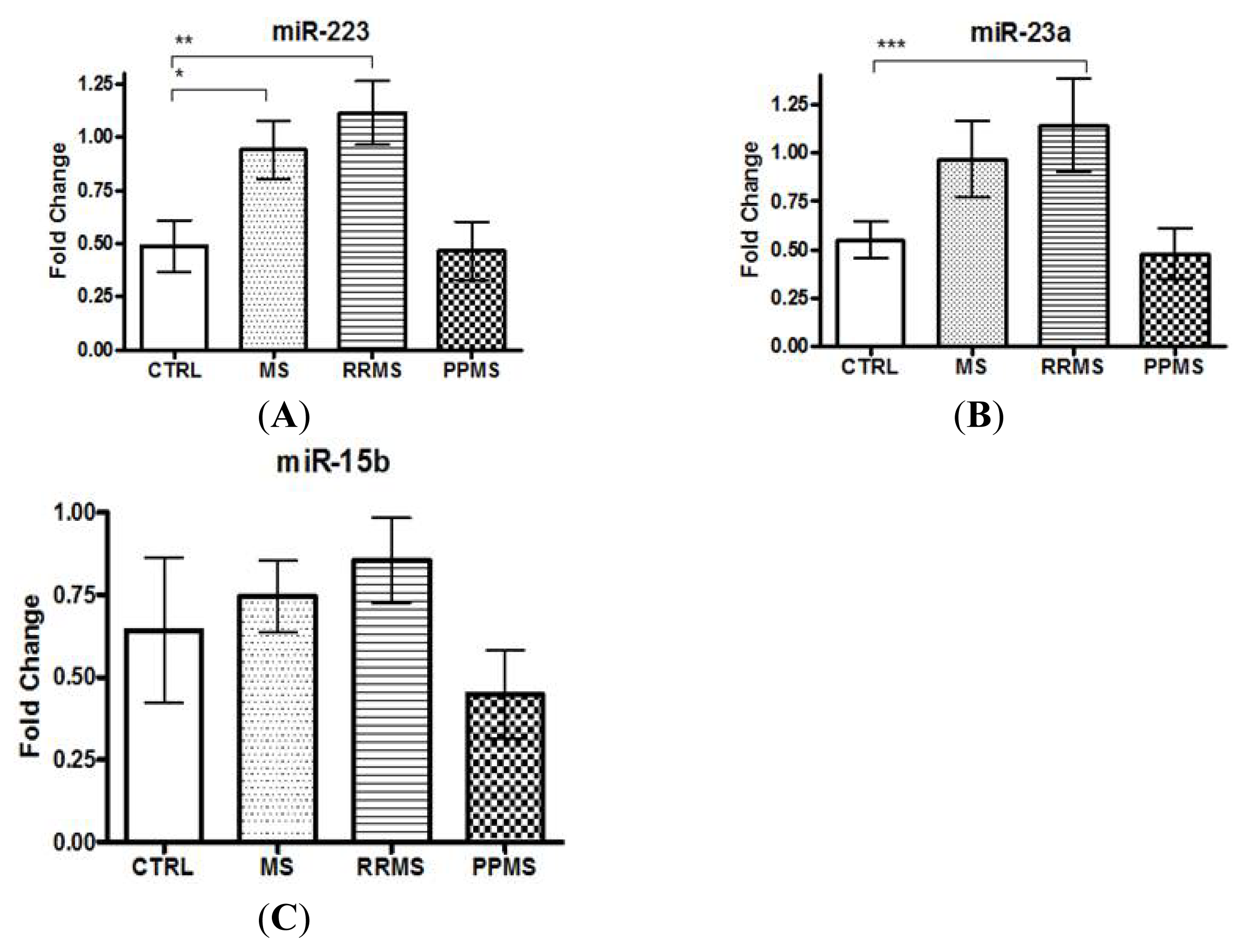
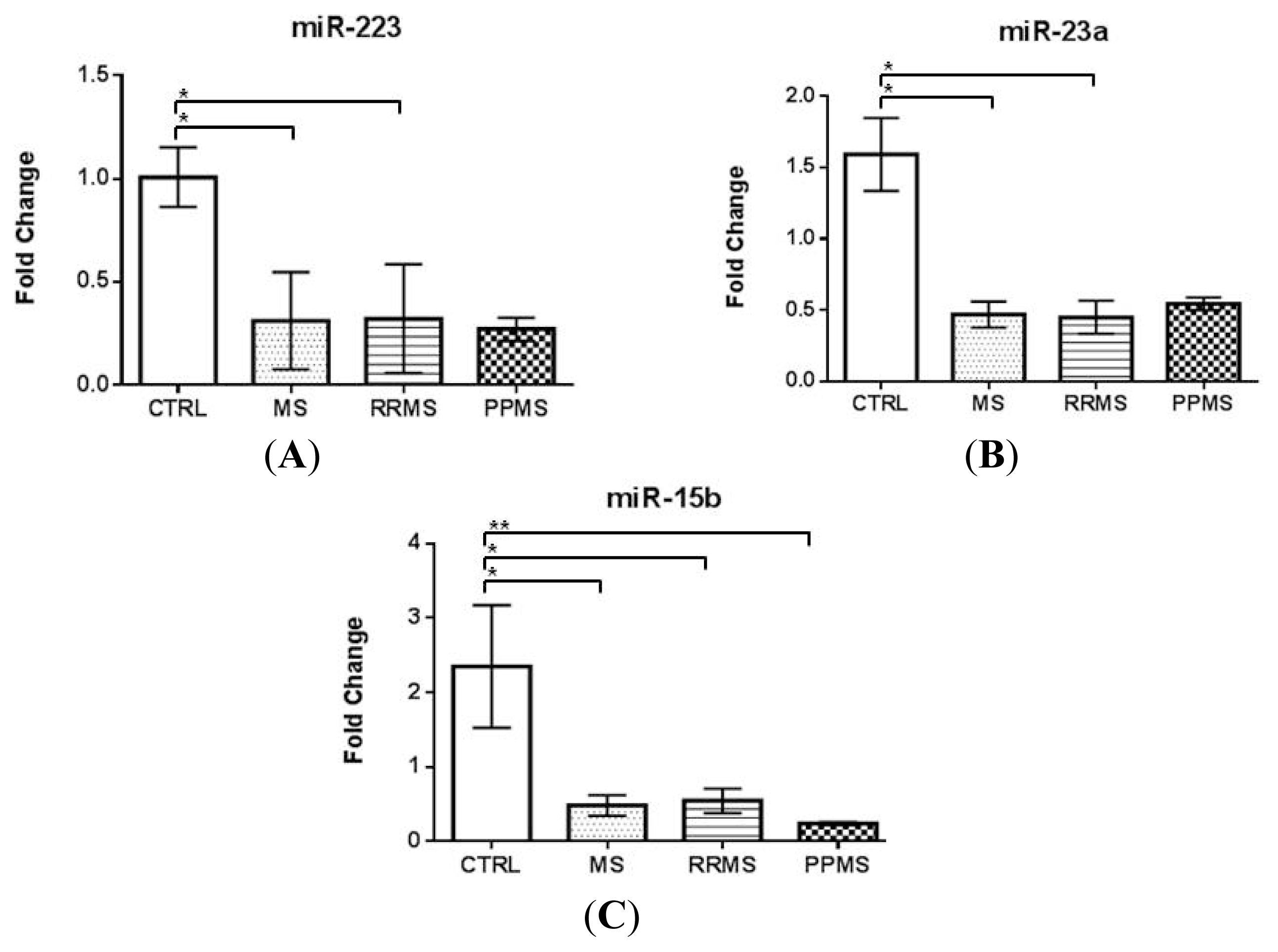
| miRNAs expression analysis population | CON | MS-all | RRMS | PPMS |
|---|---|---|---|---|
| N | 12 | 15 | 11 | 4 |
| Gender (M:F) | 5:7 | 4:11 | 2:9 | 2:2 |
| Mean age, years ± SEM | 33.9 ± 2.4 | 40.3 ± 3.2 | 36.2 ± 2.8 | 51.8 ± 6.7 |
| Mean age at onset, years ± SEM | 35.7 ± 2.6 | 31.4 ± 2.0 | 45.5 ± 4.6 | |
| Mean disease duration, years ± SEM | 3.5 ± 1.3 | 2.1 ± 0.8 | 6.8 ± 3.5 |
| SNP | n° | Genotype n (%) | Allele n (%) | |||
|---|---|---|---|---|---|---|
| miR-223 | ||||||
| rs1044165 | CC | CT | TT | C | T | |
| Controls | 420 | 348 (82.9) | 13 (4.3) | 59 (14.0) | 709 (84.4) | 131 (15.6) |
| MS patients | 399 | 305 (76.4) | 76 (19.0) | 18 (4.5) * | 686 (86.0) | 112 (14.0) |
| miR-23a | ||||||
| rs3745453 | ||||||
| Controls | 420 | 240 (57.1) | 159 (37.9) | 21 (5.0) | 639 (76.1) | 201 (23.9) |
| MS patients | 399 | 176 (44.1) | 168 (42.1) | 55 (13.8) | 520 (65.2) | 278 (34.8) ** |
| miR-15b | ||||||
| rs1451761 | ||||||
| Controls | 420 | 121 (28.8) | 201 (47.9) | 98 (23.3) | 443 (52.7) | 397 (47.3) |
| MS patients | 399 | 117 (29.3) | 182 (45.6) | 100 (25.1) | 416 (49.5) | 382 (48.9) |
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ridolfi, E.; Fenoglio, C.; Cantoni, C.; Calvi, A.; De Riz, M.; Pietroboni, A.; Villa, C.; Serpente, M.; Bonsi, R.; Vercellino, M.; et al. Expression and Genetic Analysis of MicroRNAs Involved in Multiple Sclerosis. Int. J. Mol. Sci. 2013, 14, 4375-4384. https://doi.org/10.3390/ijms14034375
Ridolfi E, Fenoglio C, Cantoni C, Calvi A, De Riz M, Pietroboni A, Villa C, Serpente M, Bonsi R, Vercellino M, et al. Expression and Genetic Analysis of MicroRNAs Involved in Multiple Sclerosis. International Journal of Molecular Sciences. 2013; 14(3):4375-4384. https://doi.org/10.3390/ijms14034375
Chicago/Turabian StyleRidolfi, Elisa, Chiara Fenoglio, Claudia Cantoni, Alberto Calvi, Milena De Riz, Anna Pietroboni, Chiara Villa, Maria Serpente, Rossana Bonsi, Marco Vercellino, and et al. 2013. "Expression and Genetic Analysis of MicroRNAs Involved in Multiple Sclerosis" International Journal of Molecular Sciences 14, no. 3: 4375-4384. https://doi.org/10.3390/ijms14034375




