Nuclear Lipid Microdomain as Place of Interaction between Sphingomyelin and DNA during Liver Regeneration
Abstract
:1. Introduction
2. Results and Discussion
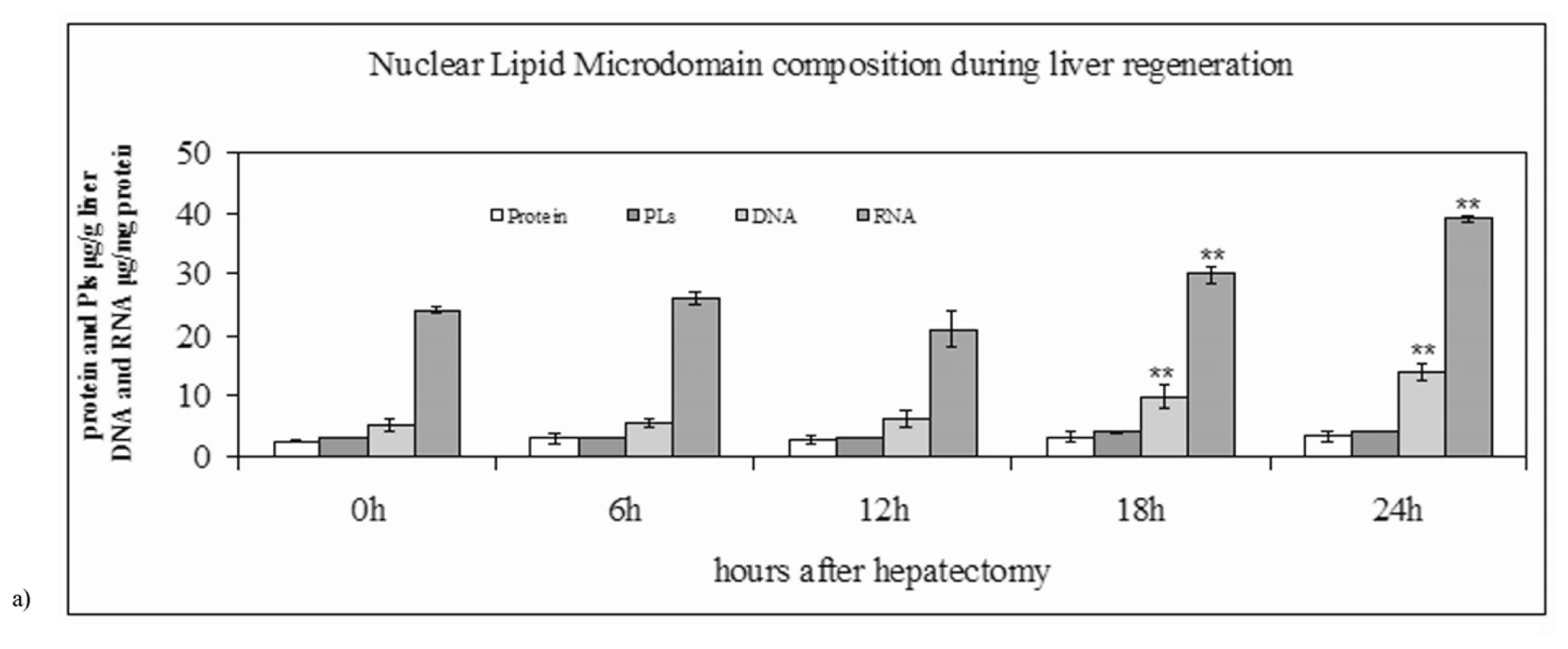
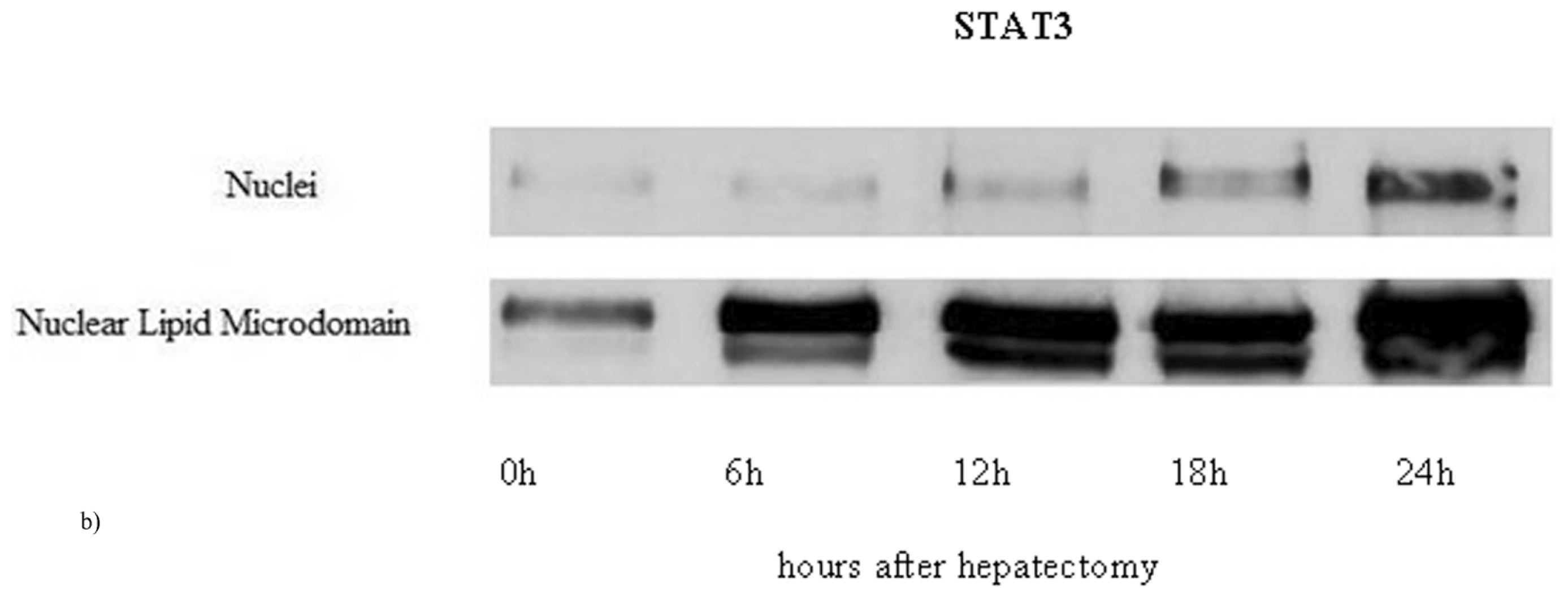
2.1. Nuclear Lipid Microdomains during Liver Regeneration
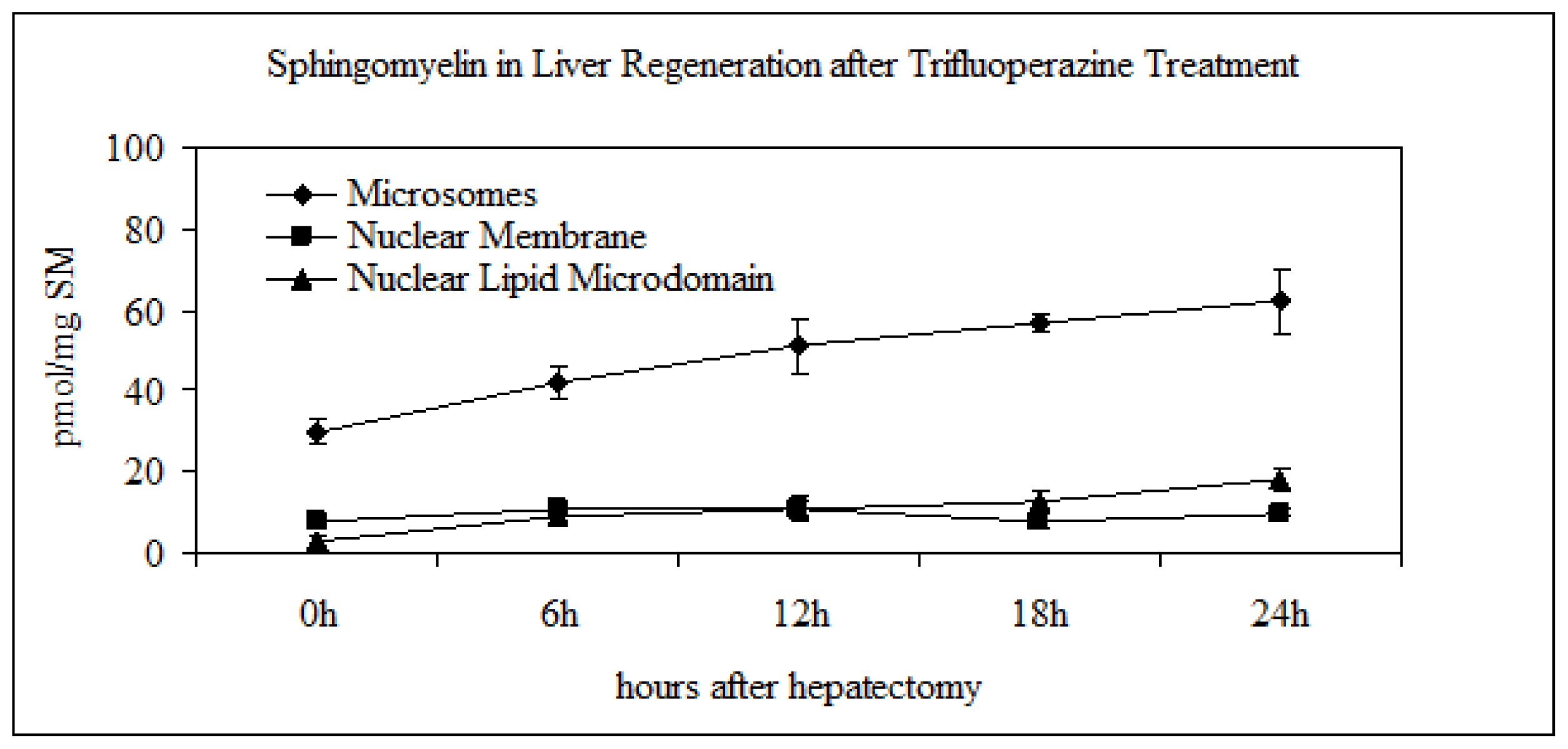
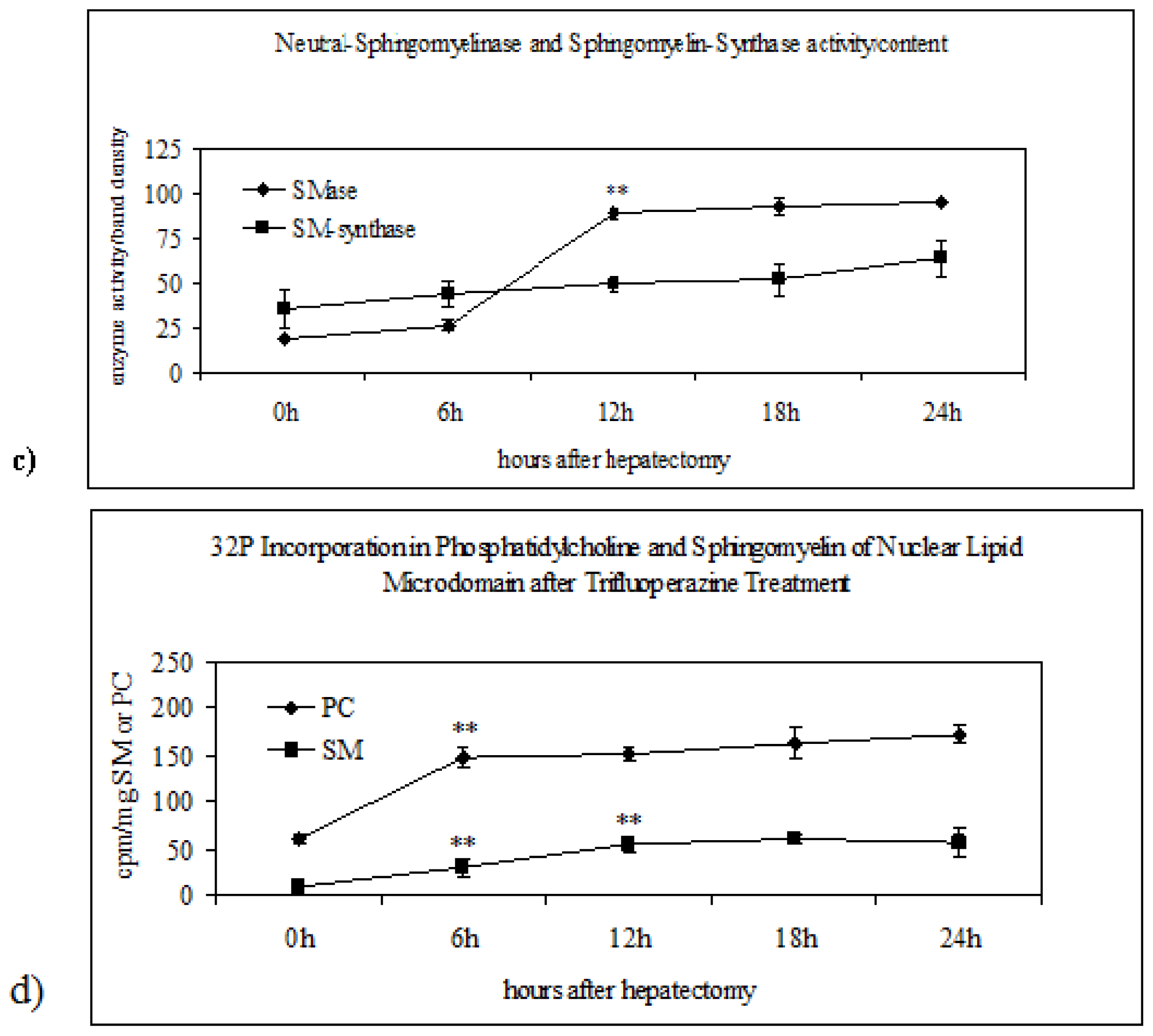
2.2. Sphingomyelin in Nuclear Lipid Microdomain during Liver Regeneration
2.3. Effect of Trifluoroperazine Treatment
3. Experimental Section
3.1. Materials
3.2. Animals
3.3. Liver Regeneration
3.4. Microsomes and Subnuclear Fractions Purification
3.5. DNA Synthesis
3.6. Lipid Analysis
3.7. Sphingomyelinase Treatment
3.8. Neutral-Sphingomyelinase and Sphingomyelin-Synthase Activity
3.9. Electrophoresis and Western Blot Analysis
4. Conclusions
Acknowledgements
Conflict of Interest
References
- Pusztai, L.; Lewis, C.E.; Yap, E. Cell proliferation in cancer. Oxford Med. Pub. 1996, 25–44. [Google Scholar]
- Xu, C.; Lin, F.; Qin, S. Relevance between lipid metabolism-associated genes and rat liver regeneration. Hepatol. Res 2008, 38, 825–837. [Google Scholar]
- Albi, E.; Viola Magni, M.P. The presence and the role of chromatin cholesterol in rat liver regeneration. J. Hepatol 2002, 36, 395–400. [Google Scholar]
- Bucher, N.L.; Weir, G.C. Insulin, glucagon, liver regeneration, and DNA synthesis. Metabolism 1976, 25, 1423–1425. [Google Scholar]
- Fausto, N.; Campbell, J.S.; Riehle, K.J. Liver regeneration. Hepatology 2006, 43, 45–53. [Google Scholar]
- Albi, E.; Magni, M.P. Chromatin neutral sphingomyelinase and its role in hepatic regeneration. Biochem. Biophys. Res. Commun 1997, 236, 29–33. [Google Scholar]
- Albi, E.; Rossi, G.; Maraldi, N.M.; Magni, M.V.; Cataldi, S.; Solimando, L.; Zini, N. Involvement of nuclear phosphatidylinositol-dependent phospholipases C in cell cycle progression during rat liver regeneration. J. Cell Physiol 2003, 197, 181–188. [Google Scholar]
- Albi, E. The role of intranuclear lipids in health and disease. Clin. Lipidol 2011, 6, 59–69. [Google Scholar]
- Damaskopoulou, E.; Keri, G.; Jacob, V.; Lazzarini, R.; Cataldi, S.; Orfi, L.; Albi, E. Protein kinase Cζ drives Sphingomyelin Metabolism in the Nucleus during Cell Proliferation. Curr. Chem. Biol. 2013, in press. [Google Scholar]
- Viola Magni, M.P.; Gahan, P.B.; Albi, E.; Iapoce, R.; Gentilucci, P.F. Chromatin phospholipids and DNA synthesis in hepatic cells. Basic Appl. Histochem 1985, 29, 253–259. [Google Scholar]
- Zabielski, P.; Baranowski, M.; Zendzian-Piotrowska, M.; Blachnio, A.; Gorski, J. Partial hepatectomy activates production of the pro-mitotic intermediates of the sphingomyelin signal transduction pathway in the rat liver. Prostaglandins Other Lipid Mediat 2007, 83, 277–284. [Google Scholar]
- Zabielski, P.; Baranowski, M.; Zendzian-Piotrowska, M.; Błachnio-Zabielska, A.; Górski, J. Bezafibrate decreases growth stimulatory action of the sphingomyelin signaling pathway in regenerating rat liver. Prostaglandins Other Lipid Mediat 2008, 85, 17–25. [Google Scholar]
- Alessenko, A.V.; Platonova, L.V.; Sakevarashvili, G.R.; Khrenov, A.V.; Shingarova, L.N.; Shono, N.I.; Galperin, E.I. Role of endogenous TNF-alpha and sphingosine in induced DNA synthesis in regenerating rat liver after partial hepatectomy. Biochemistry 1999, 64, 890–895. [Google Scholar]
- Albi, E.; Viola Magni, M. The role of intranuclear lipids. Biol. Cell 2004, 96, 657–667. [Google Scholar]
- Cascianelli, G.; Villani, M.; Tosti, M.; Marini, F.; Bartoccini, E.; Viola Magni, M.; Albi, E. Lipid microdomains in cell nucleus. Mol. Biol. Cell 2008, 19, 5289–5295. [Google Scholar]
- Albi, E.; Villani, M. Nuclear lipid microdomains regulate cell function. Commun. Integr. Biol 2009, 2, 23–24. [Google Scholar]
- Ratnakar, P.; Murthy, P.S. Trifluoperazine inhibits the incorporation of labelled precursors into lipids, proteins and DNA of Mycobacterium tuberculosis H37Rv. FEMS Microbiol. Lett 1993, 110, 291–294. [Google Scholar]
- Sharma, S.; Kaur, H.; Khuller, G.K. Cell cycle effects of the phenothiazines: trifluoperazine and chlorpromazine in Candida albicans. FEMS Microbiol. Lett 2001, 199, 185–190. [Google Scholar]
- Soriano, M.; Piñol, R.; Enrich, C.; Bachs, O. Effect of trifluoperazine on DNA synthesis during liver regeneration. Cell Tissue Kinet 1985, 18, 475–481. [Google Scholar]
- Kolesnick, R.N.; Hemer, M.R. Trifluoperazine stimulates the coordinate degradation of sphingomyelin and phosphatidylcholine in GH3 pituitary cells. J. Biol. Chem 1989, 264, 14057–14061. [Google Scholar]
- Rossi, G.; Viola Magni, M.; Albi, E. Signal transducer and activator of transcription 3 and sphingomyelin metabolism in intranuclear complex during cell proliferation. Arch. Biochem. Biophys 2007, 464, 138–143. [Google Scholar]
- Ledeen, R.W.; Wu, G. Sphingolipids of the nucleus and their role in nuclear signalling. Biochim. Biophys. Acta 2006, 1761, 588–598. [Google Scholar]
- Albi, E.; Viola Magni, M. Sphingomyelin: A Small-Big Molecule in the Nucleus. In Sphingolipid and Cell Function; Albi, E., Ed.; Research Signpost: Kerale, India, 2006; Volume 37, pp. 211–227. [Google Scholar]
- Tomassoni, M.L.; Amori, D.; Magni, M.V. Changes of nuclear membrane lipid composition affect RNA nucleocytoplasmic transport. Biochem. Biophys. Res. Commun 1999, 258, 476–481. [Google Scholar]
- Scassellati, C.; Albi, E.; Cmarko, D.; Tiberi, C.; Cmarkova, J.; Bouchet-Marquis, C.; Verschure, P.J.; Driel, R.; Magni, M.V.; Fakan, S. Intranuclear sphingomyelin is associated with transcriptionally active chromatin and plays a role in nuclear integrity. Biol. Cell 2010, 102, 361–375. [Google Scholar]
- Viola Magni, M.P.; Gahan, P.B.; Albi, E.; Iapoce, R.; Gentilucci, P.F. Phospholipids in chromatin: incorporation of [32P]O4 2− in different subcellular fraction of hepatocytes. Cell Biochem. Funct 1986, 4, 283–288. [Google Scholar]
- Kay, R.R.; Johnston, I.R. Rapid isolation of nuclear envelopes from rat liver. Methods Cell Biol 1975, 15, 277–283. [Google Scholar]
- Folch, J.; Lees, M.; Sloene-Stanley, G.H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem 1957, 226, 497–509. [Google Scholar]
- Bartlett, G.R. Phosphorus assay in column chromatography. J. Biol. Chem 1959, 234, 466–468. [Google Scholar]
- Christie, W.W. Lipid Analysis: Isolation, Separation, Identification and Structural Analysis of Lipids; Pergamon Press: Oxford, UK, 1973; pp. 152–325. [Google Scholar]
- Albi, E.; Lazzarini, R.; Viola Magni, M. Phosphatidylcholine/sphingomyelin metabolism crosstalk inside the nucleus. Biochem. J 2008, 410, 381–389. [Google Scholar]





| Time | Nuclear membrane | Nuclear matrix | Chromatin | Nuclear microdomain |
|---|---|---|---|---|
| 0 h | 9.3 ± 0.7 | 1.6 ± 0.3 | 0.9 ± 0.1 | 15.5 ± 0.3 |
| 6 h | 12.8 ± 0.5 * | 3.7 ± 0.6 * | 0.7 ± 0.2 | 14.1 ± 0.2 |
| 12 h | 12.1 ± 1.3 * | 6.5 ± 0.5 * | 2.2 ± 0.1 * | 38.2 ± 0.7 * |
| 18 h | 21.3 ± 0.7 * | 7.7 ± 0.2 * | 0.9 ± 0.1 | 15.7 ± 0.1 |
| 24 h | 9.3 ± 0.3 | 6.8 ± 0.5 * | 1.2 ± 0.2 | 19.5 ± 0.2 * |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Albi, E.; Lazzarini, A.; Lazzarini, R.; Floridi, A.; Damaskopoulou, E.; Curcio, F.; Cataldi, S. Nuclear Lipid Microdomain as Place of Interaction between Sphingomyelin and DNA during Liver Regeneration. Int. J. Mol. Sci. 2013, 14, 6529-6541. https://doi.org/10.3390/ijms14046529
Albi E, Lazzarini A, Lazzarini R, Floridi A, Damaskopoulou E, Curcio F, Cataldi S. Nuclear Lipid Microdomain as Place of Interaction between Sphingomyelin and DNA during Liver Regeneration. International Journal of Molecular Sciences. 2013; 14(4):6529-6541. https://doi.org/10.3390/ijms14046529
Chicago/Turabian StyleAlbi, Elisabetta, Andrea Lazzarini, Remo Lazzarini, Alessandro Floridi, Eleni Damaskopoulou, Francesco Curcio, and Samuela Cataldi. 2013. "Nuclear Lipid Microdomain as Place of Interaction between Sphingomyelin and DNA during Liver Regeneration" International Journal of Molecular Sciences 14, no. 4: 6529-6541. https://doi.org/10.3390/ijms14046529





