Regulatory Roles of MicroRNAs in Diabetes
Abstract
:1. MicroRNAs (miRNAs) and Diabetes
2. miRNAs and Pancreatic β-Cells
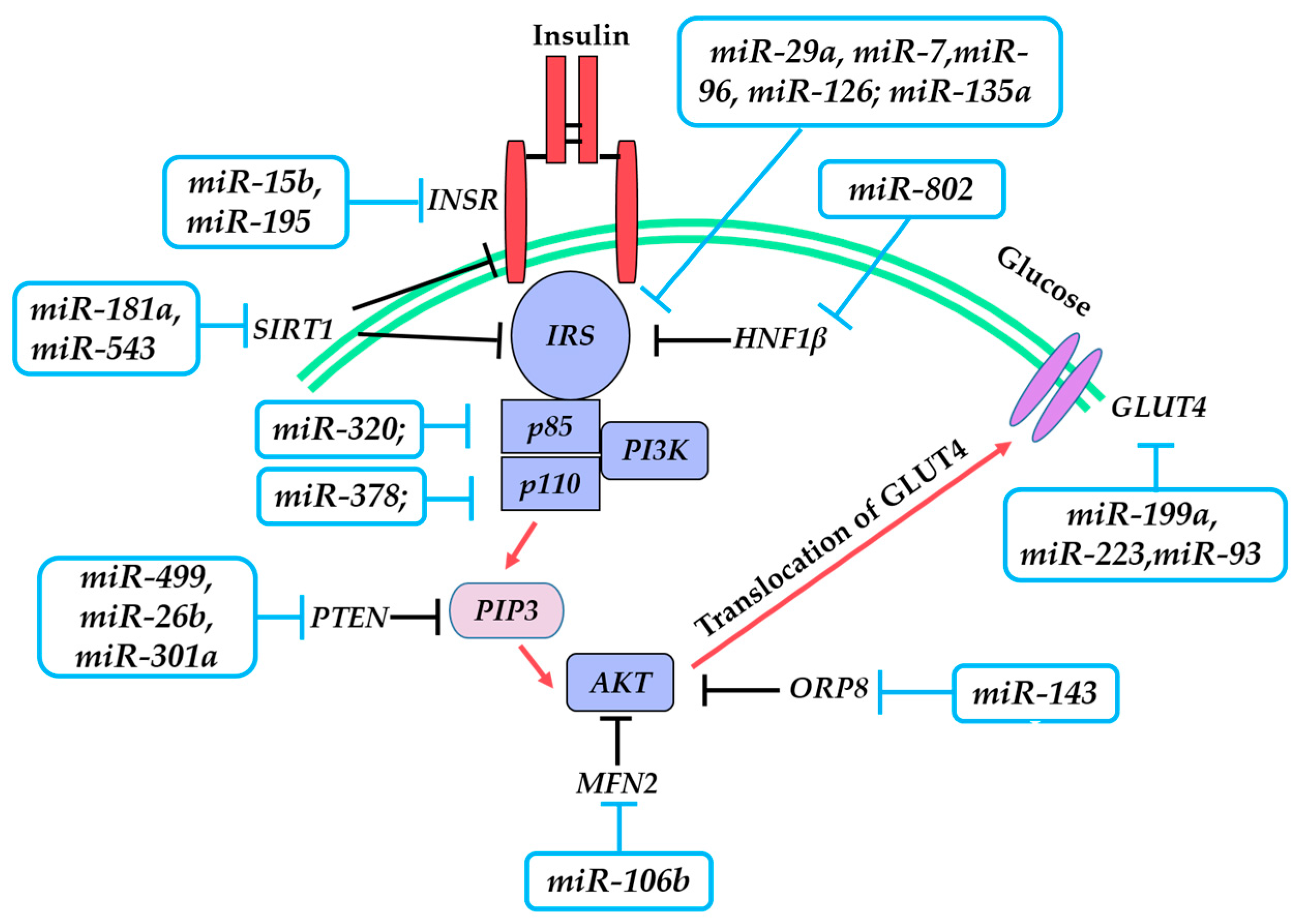
3. miRNAs and Insulin Resistance (IR)
3.1. Insulin Receptor (INSR)
3.2. Insulin Receptor Substrate 1/2 (IRS-1/2)
3.3. Phosphoinositide 3-Kinase (PI3K)/AKT Serine/Threonine Kinase (AKT)
3.4. Glucose Transporter 4 (GLUT4)
4. Systematic Analysis of miRNA–mRNA Regulatory Network in Diabetes
5. miRNAs as Type 2 Diabetes (T2DM) Markers and Its Complications
6. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of interest
Appendix A
| Gene Symbol Abbreviations | Gene’s Full Names |
|---|---|
| Bcl-2 | B-cell lymphoma-2 |
| Trp53 | transformation related protein 53 |
| Bax | BCL2 associated X, apoptosis regulator |
| Cadm1 | cell adhesion molecule 1 |
| Hnf1α | hepatocyte nuclear factor 1 homeobox A |
| Neurod1 | neurogenic differentiation 1 |
| HNF1β | hepatocyte nuclear factor 1 homeobox β |
| PAX6 | paired box 6 |
| Stx-1 | syntaxin 1 |
| Rab27a | Ras-related protein Rab-27A |
| Foxa2 | forkhead box protein A2 |
| Fgf21 | fibroblast growth factor 21 |
| Pdgfrα | platelet-derived growth factor receptor, α polypeptide |
| RFX6 | regulatory factor X6 |
| SOX6 | sex determining region Y-box 6 |
| Mtpn | myotrophin |
| Ago2 | argonaute 2 |
| Mct1 | monocarboxylate transporter 1 |
| HIPK3 | homeodomain interacting protein kinase 3 |
| Abca1 | ATP binding cassette subfamily A member 1 |
| INSR | insulin receptor |
| IRS-1 | insulin receptor substrate-1 |
| IRS-2 | insulin receptor substrate-2 |
| PI3K | phosphatidylinositol 3-kinase |
| GLUT4 | glucose transporter member 4 |
| PTEN | phosphatase and tensin homolog |
| Orp8 | oxysterol binding protein like 8 |
| SIRT-1 | sirtuin 1 |
| Mfn2 | mitofusin 2 |
| ADAM22 | ADAM metallopeptidase domain 22 |
| MYO5A | myosin VA |
| LOX | lysyl oxidase |
| GM2A | GM2 ganglioside activator |
| ZEB2 | zinc finger E-box binding homeobox 2 |
| TGF-β | transforming growth factor β |
| SPRY1 | sprouty homolog 1 |
| THBS-1 | thrombospondin 1 |
References
- Olokoba, A.B.; Obateru, O.A.; Olokoba, L.B. Type 2 diabetes mellitus: A review of current trends. Oman Med. J. 2012, 27, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M. History of diabetes mellitus. Saudi Med. J. 2002, 23, 373–378. [Google Scholar] [PubMed]
- Raghunathan, K. History of diabetes from remote to recent times. Bull. Indian Inst. Hist. Med. Hyderabad 1976, 6, 167–182. [Google Scholar] [PubMed]
- Mark, A.A.; George, S.E.; Aaron, M. Type 1 diabetes. Lancet 2014, 383, 69–82. [Google Scholar]
- Chen, L.; Magliano, D.J.; Zimmet, P.Z. The worldwide epidemiology of type 2 diabetes mellitus—Present and future perspectives. Nat. Rev. Endocrinol. 2011, 8, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Sekar, D.; Venugopal, B.; Sekar, P.; Ramalingam, K. Role of microRNA 21 in diabetes and associated/related diseases. Gene 2016, 582, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification of MIRNA genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Dey, N.; Das, F.; Mariappan, M.M.; Mandal, C.C.; Ghosh-Choudhury, N.; Kasinath, B.S.; Choudhury, G.G. MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes. J. Biol. Chem. 2011, 286, 25586–25603. [Google Scholar] [CrossRef] [PubMed]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Melkman-Zehavi, T.; Oren, R.; Kredo-Russo, S.; Shapira, T.; Mandelbaum, A.D.; Rivkin, N.; Nir, T.; Lennox, K.A.; Behlke, M.A.; Dor, Y.; et al. MiRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. EMBO J. 2011, 30, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Li, G.M.; Dong, Q.; Peng, H. MiR-577 inhibits pancreatic β-cell function and survival by targeting fibroblast growth factor 21 (FGF-21) in pediatric diabetes. Genet. Mol. Res. 2015, 14, 15462–15470. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, Z.; Tu, Y.; Shen, H.; Dai, Z.; Lin, J.; Zhou, Z. miR-101a and miR-30b contribute to inflammatory cytokine-mediated β-cell dysfunction. Lab. Investig. 2015, 95, 1387–1397. [Google Scholar] [CrossRef] [PubMed]
- Belgardt, B.F.; Ahmed, K.; Spranger, M.; Latreille, M.; Denzler, R.; Kondratiuk, N.; von Meyenn, F.; Villena, F.N.; Herrmanns, K.; Bosco, D.; et al. The microRNA-200 family regulates pancreatic β-cell survival in type 2 diabetes. Nat. Med. 2015, 21, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Guan, H.; Huang, Z.; Liu, J.; Li, H.; Wei, G.; Cao, X.; Li, Y. Downregulation of Bcl-2 expression by miR-34a mediates palmitate-induced Min6 cells apoptosis. J. Diabetes Res. 2014, 258695. [Google Scholar] [CrossRef] [PubMed]
- Poy, M.N.; Hausser, J.; Trajkovski, M.; Braun, M.; Collins, S.; Rorsman, P.; Zavolan, M.; Stoffel, M. MiR-375 maintains normal pancreatic α- and β-cell mass. Proc. Natl. Acad. Sci. USA 2009, 106, 5813–5818. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xie, H.Y.; Ding, S.M.; Xing, C.Y.; Chen, A.; Lai, M.C.; Zhou, L.; Zheng, S.S. CADM1 regulates the G1/S transition and represses tumorigenicity through the Rb-E2F pathway in hepatocellular carcinoma. Hepatob. Pancreat. Dis. Int. 2016, 15, 289–296. [Google Scholar] [CrossRef]
- Lu, Y.; Fei, X.Q.; Yang, S.F.; Xu, B.K.; Li, Y.Y. Glucose-induced microRNA-17 promotes pancreatic β-cell proliferation through down-regulation of Menin. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 624–629. [Google Scholar] [PubMed]
- Chen, Y.; Tian, L.; Wan, S.; Xie, Y.; Chen, X.; Ji, X.; Zhao, Q.; Wang, C.; Zhang, K.; Hock, J.M.; et al. MicroRNA-17-92 cluster regulates pancreatic β-cell proliferation and adaptation. Mol. Cell. Endocrinol. 2016, 437, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; You, W.; Wang, H.; Li, Y.; Qiao, N.; Shi, Y.; Zhang, C.; Bleich, D.; Han, X. MicroRNA-24/MODY gene regulatory pathway mediates pancreatic β-cell dysfunction. Diabetes 2013, 62, 3194–3206. [Google Scholar] [CrossRef] [PubMed]
- Bagge, A.; Clausen, T.R.; Larsen, S.; Ladefoged, M.; Rosenstierne, M.W.; Larsen, L.; Vang, O.; Nielsen, J.H.; Dalgaard, L.T. MicroRNA-29a is up-regulated in β-cells by glucose and decreases glucose-stimulated insulin secretion. Biochem. Biophys. Res. Commun. 2012, 426, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Yang, J.; Liu, G.Q.; Gao, M.J.; Hou, W.F.; Zhang, L.; Gao, H.W.; Liu, Y.; Chen, G.A.; Hong, T.P. Dynamic expression of microRNAs during the differentiation of human embryonic stem cells into insulin-producing cells. Gene 2013, 518, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Kredo-Russo, S.; Mandelbaum, A.D.; Ness, A.; Alon, I.; Lennox, K.A.; Behlke, M.A.; Hornstein, E. Pancreas-enriched miRNA refines endocrine cell differentiation. Development 2012, 139, 3021–3031. [Google Scholar] [CrossRef] [PubMed]
- Shae, A.; Azarpira, N.; Karimi, M.H.; Soleimani, M.; Dehghan, S. Differentiation of human-induced pluripotent stem cells into insulin-producing clusters by microRNA-7. Exp. Clin. Transplant. 2015, 16, 121–128. [Google Scholar]
- Nathan, G.; Kredo-Russo, S.; Geiger, T.; Lenz, A.; Kaspi, H.; Hornstein, E.; Efrat, S. MiR-375 promotes redifferentiation of adult human β-cells expanded in vitro. PLoS ONE 2015, 10, e0122108. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Xue, H.; Wang, Y.C.; Nazor, K.L.; Guo, S.; Trivedi, N.; Peterson, S.E.; Liu, Y.; Loring, J.F.; Laurent, L.C. Matched miRNA and mRNA signatures from an hESC-based in vitro model of pancreatic differentiation reveal novel regulatory interactions. J. Cell Sci. 2013, 126, 3848–3861. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Li, X.; Gao, Y.; Wang, K.; Fan, Y.; Zhang, S.; Ma, Y.; Guan, W. Role of microRNA-21 in the formation of insulin-producing cells from pancreatic progenitor cells. Biochim. Biophys. Acta 2016, 1859, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, M.V.; Joglekar, V.M.; Hardikar, A.A. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr. Patterns 2009, 9, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Poy, M.N.; Eliasson, L.; Krutzfeldt, J.; Kuwajima, S.; Ma, X.; Macdonald, P.E.; Pfeffer, S.; Tuschl, T.; Rajewsky, N.; Rorsman, P.; et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Tattikota, S.G.; Rathjen, T.; Hausser, J.; Khedkar, A.; Kabra, U.D.; Pandey, V.; Sury, M.; Wessels, H.H.; Mollet, I.G.; Eliasson, L.; et al. MiR-184 regulates pancreatic β-cell function according to glucose metabolism. J. Biol. Chem. 2015, 290, 20284–20294. [Google Scholar] [CrossRef] [PubMed]
- Latreille, M.; Hausser, J.; Stützer, I.; Zhang, Q.; Hastoy, B.; Gargani, S.; Kerr-Conte, J.; Pattou, F.; Zavolan, M.; Esguerra, J.L.; et al. MicroRNA-7a regulates pancreatic β-cell function. J. Clin. Investig. 2014, 124, 2722–2735. [Google Scholar] [CrossRef] [PubMed]
- Gomes, P.R.; Graciano, M.F.; Pantaleão, L.C.; Rennó, A.L.; Rodrigues, S.C.; Velloso, L.A.; Latorraca, M.Q.; Carpinelli, A.R.; Anhê, G.F.; Bordin, S. Long-term disruption of maternal glucose homeostasis induced by prenatal glucocorticoid treatment correlates with miR-29 upregulation. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E109–E120. [Google Scholar] [CrossRef] [PubMed]
- Pullen, T.J.; da Silva Xavier, G.; Kelsey, G.; Rutter, G.A. MiR-29a and miR-29b contribute to pancreatic β-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol. Cell. Biol. 2011, 31, 3182–3194. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.M.; da Silva Xavier, G.; Dawe, H.R.; Rutter, G.A.; Harries, L.W. Increased expression of miR-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia 2014, 57, 122–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.W.; You, Y.H.; Jung, S.; Suh-Kim, H.; Lee, I.K.; Cho, J.H.; Yoon, K.H. MiRNA-30a-5p-mediated silencing of β2/NeuroD expression is an important initial event of glucotoxicity-induced β-cell dysfunction in rodent models. Diabetologia 2013, 56, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, G.; Po, A.; Miele, E.; Ventriglia, G.; Ceccarelli, E.; Bugliani, M.; Marselli, L.; Marchetti, P.; Gulino, A.; Ferretti, E.; et al. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol. 2015, 52, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Baroukh, N.; Ravier, M.A.; Loder, M.K.; Hill, E.V.; Bounacer, A.; Scharfmann, R.; Rutter, G.A.; Van Obberghen, E. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic β-cell lines. J. Biol. Chem. 2007, 282, 19575–19588. [Google Scholar] [CrossRef] [PubMed]
- Wijesekara, N.; Zhang, L.H.; Kang, M.H.; Abraham, T.; Bhattacharjee, A.; Warnock, G.L.; Verchere, C.B.; Hayden, M.R. MiR-33a modulates ABCA1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes 2012, 61, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Stumvoll, M.; Goldstein, B.J.; van Haeften, T.W. Type 2 diabetes: Principles of pathogenesis and therapy. Lancet 2005, 365, 1333–1346. [Google Scholar] [CrossRef]
- Yang, W.M.; Jeong, H.J.; Park, S.Y.; Lee, W. Saturated fatty acid-induced miR-195 impairs insulin signaling and glycogen metabolism in HepG2 cells. FEBS Lett. 2014, 588, 3939–3946. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.M.; Jeong, H.J.; Park, S.W.; Lee, W. Obesity-induced miR-15b is linked causally to the development of insulin resistance through the repression of the insulin receptor in hepatocytes. Mol. Nutr. Food Res. 2015, 59, 2303–2314. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.M.; Jeong, H.J.; Park, S.Y.; Lee, W. Induction of miR-29a by saturated fatty acids impairs insulin signaling and glucose uptake through translational repression of IRS-1 in myocytes. FEBS Lett. 2014, 588, 2170–2176. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Na, H.M.; Peng, G.; Pu, J.; Liu, P. Alteration of microRNA expression correlates to fatty acid-mediated insulin resistance in mouse myoblasts. Mol. Biosyst. 2011, 7, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Park, S.Y.; Yang, W.M.; Lee, W. The induction of miR-96 by mitochondrial dysfunction causes impaired glycogen synthesis through translational repression of IRS-1 in SK-Hep1 cells. Biochem. Biophys. Res. Commun. 2013, 434, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Twinn, D.S.; Alfaradhi, M.Z.; Martin-Gronert, M.S.; Duque-Guimaraes, D.E.; Piekarz, A.; Ferland-McCollough, D.; Bushell, M.; Ozanne, S.E. Downregulation of IRS-1 in adipose tissue of offspring of obese mice is programmed cell-autonomously through post-transcriptional mechanisms. Mol. Metab. 2014, 3, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Srivastava, R.; Srivastava, A.K.; Ali, S.; Datta, M. MiR-135a targets IRS2 and regulates insulin signaling and glucose uptake in the diabetic gastrocnemius skeletal muscle. Biochim. Biophys. Acta 2013, 1832, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Cao, H.; Ye, C.; Chang, C.; Lu, M.; Jing, Y.; Zhang, D.; Yao, X.; Duan, Z.; Xia, H.; et al. Hepatic miR-378 targets p110α and controls glucose and lipid homeostasis by modulating hepatic insulin signalling. Nat. Commun. 2014, 5, 5684–5695. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.Y.; Ou, H.S.; Feng, S.D.; Zhang, X.Y.; Tuo, Q.H.; Chen, L.X.; Zhu, B.Y.; Gao, Z.P.; Tang, C.K.; Yin, W.D.; et al. Changes in microRNA (miR) profile and effects of miR-320 in insulin-resistant 3T3-L1 adipocytes. Clin. Exp. Pharmacol. Physiol. 2009, 36, e32–e39. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, N.; Pan, H.P.; Wang, Z.; Cao, Z.Y. MiR-499–5p contributes to hepatic insulin resistance by suppressing PTEN. Cell. Physiol. Biochem. 2015, 36, 2357–2365. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Ji, C.; Song, G.; Zhao, C.; Shi, C.; Song, L.; Chen, L.; Yang, L.; Huang, F.; Pang, L.; Zhang, N.; Zhao, Y.; Guo, X. MiR-26b modulates insulin sensitivity in adipocytes by interrupting the PTEN/PI3K/AKT pathway. Int. J. Obes. 2015, 39, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Wang, S.; Sui, X.; Meng, X.; Shen, T.; Huang, X.; Guo, J.; Fang, W.; Man, Y.; Xi, J.; et al. MiR-301a mediates the effect of IL-6 on the AKT/GSK pathway and hepatic glycogenesis by regulating PTEN expression. Cell. Physiol. Biochem. 2015, 35, 1413–1424. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.T.; Li, C.L.; Tian, H.; Li, J.; Pei, Y.; Liu, Y.; Gong, Y.P.; Fang, F.S.; Sun, B.R. MiR-199a is overexpressed in plasma of type 2 diabetes patients which contributes to type 2 diabetes by targeting GLUT4. Mol. Cell. Biochem. 2014, 397, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.Y.; Wu, H.L.; Chen, C.C.; Gamboa, G.M.; Layman, L.C.; Diamond, M.P.; Azziz, R.; Chen, Y.H. MicroRNA-223 expression is upregulated in insulin resistant human adipose tissue. J. Diabetes Res. 2015, 2015, 943659–943709. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Heneidi, S.; Lee, J.M.; Layman, L.C.; Stepp, D.W.; Gamboa, G.M.; Chen, B.S.; Chazenbalk, G.; Azziz, R. MiRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes 2013, 62, 2278–2286. [Google Scholar] [CrossRef] [PubMed]
- Blumensatt, M.; Greulich, S.; Herzfeld de Wiza, D.; Mueller, H.; Maxhera, B.; Rabelink, M.J.; Hoeben, R.C.; Akhyari, P.; Al-Hasani, H.; Ruige, J.B.; et al. Activin A impairs insulin action in cardiomyocytes via up-regulation of miR-143. Cardiovasc. Res. 2013, 100, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Jordan, S.D.; Krüger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Brönneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Böttger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 2011, 13, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Kornfeld, J.W.; Baitzel, C.; Könner, A.C.; Nicholls, H.T.; Vogt, M.C.; Herrmanns, K.; Scheja, L.; Haumaitre, C.; Wolf, A.M.; Knippschild, U.; et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature 2013, 494, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Li, C.; Qi, W.; Zhang, Y.; Zhang, F.; Wu, J.X.; Hu, Y.N.; Wu, D.M.; Liu, Y.; Yan, T.T.; et al. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia 2012, 55, 2032–2043. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gu, P.; Shi, W.; Li, J.; Hao, Q.; Cao, X.; Lu, Q.; Zeng, Y. MicroRNA-29a induces insulin resistance by targeting PPARδ in skeletal muscle cells. Int. J. Mol. Med. 2016, 37, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chi, L.; Zhang, W.; Bai, T.; Zhao, W.; Feng, Z.; Tian, H. Down-regulation of the miR-543 alleviates insulin resistance through targeting the SIRT1. Biochem. Biophys. Res. Commun. 2015, 468, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.Q.; Wei, S.B.; Sun, Y.M.; Qin, W.Y.; Cheng, J.; Mitchelson, K.; Xie, L. Systematic profiling of mRNA and miRNA expression in the pancreatic islets of spontaneously diabetic Goto-Kakizaki rats. Mol. Med. Rep. 2015, 11, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Kirby, T.J.; Walton, R.G.; Finlin, B.; Zhu, B.; Unal, R.; Rasouli, N.; Peterson, C.A.; Kern, P.A. Integrative mRNA–microRNA analyses reveal novel interactions related to insulin sensitivity in human adipose tissue. Physiol. Genom. 2016, 48, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Lv, C.; Li, L.; Chen, S.; Liu, S.; Wang, C.; Su, B. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. BioMed Res. Int. 2013, 761617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, L.; Shang, Q.; Lv, C.; Wang, C.; Su, B. Circulating miR-126 is a potential biomarker to predict the onset of type 2 diabetes mellitus in susceptible individuals. Biochem. Biophys. Res. Commun. 2015, 463, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wang, T.; Huang, S.; Di, Y.; Huang, Y.; Liu, X.; Luo, Z.; Han, W.; An, B. Differential expression of microRNAs in plasma of patients with prediabetes and newly diagnosed type 2 diabetes. Acta Diabetol. 2016, 53, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chien, H.Y.; Chen, C.Y.; Chiu, Y.H.; Lin, Y.C.; Li, W.C. Differential microRNA profiles predict diabetic nephropathy progression in Taiwan. Int. J. Med. Sci. 2016, 13, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Yong, T.Y.; Michael, M.Z.; Gleadle, J.M. Review: The role of microRNAs in kidney disease. Nephrology 2010, 15, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Wang, Y.; Wang, W.; Chang, B.H.; Danesh, F.R. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J. Biol. Chem. 2011, 286, 11837–11848. [Google Scholar] [CrossRef] [PubMed]
- Delić, D.; Eisele, C.; Schmid, R.; Baum, P.; Wiech, F.; Gerl, M.; Zimdahl, H.; Pullen, S.S.; Urquhart, R. Urinary exosomal miRNA signature in type II diabetic nephropathy patients. PLoS ONE 2016, 11, e0150154. [Google Scholar] [CrossRef] [PubMed]
- Al-Kafaji, G.; Al-Mahroos, G.; Abdulla Al-Muhtaresh, H.; Sabry, M.A.; Abdul-Razzak, R.; Salem, A.H. Circulating endothelium-enriched microRNA-126 as a potential biomarker for coronary artery disease in type 2 diabetes mellitus patients. Biomarkers 2016, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
| Cell Processes | miRNA | Putative Targets | Cells or Tissues Studied | Species | References |
|---|---|---|---|---|---|
| β-cell survival/apoptosis | miR-577 | Fgf-21 | INS-1 cells | rat | [11] |
| miR-200a/b/c | - | islets of diabetic mice | mouse | [12] | |
| miR-34a | Bcl-2 | MIN-6 cells | mouse | [13] | |
| β-cell proliferation | miR-375 | Cadm1 | islets of miR-375 KO mice; INS-1E cells | mouse; rat | [14,15] |
| miR-181a | Pdgfrα | 3- and 12-month-old rat islets | rat | [16] | |
| miR-17 | Menin | MIN-6 cells | mouse | [17] | |
| miR-24 | Hnf1α; Neurod1 | islets of obese mice | mouse | [19] | |
| miR-29a | - | INS-1E cells | rat | [20] | |
| β-cell differentiation | miR-375 | HNF1β | hPSCs differentiated IPCs | human | [21] |
| miR-7 | PAX6 | hPSCs differentiated IPCs | human | [21] | |
| miR-34a | - | hPSCs differentiated IPCs | human | [21] | |
| miR-146a | - | hPSCs differentiated IPCs | human | [21] | |
| miR-30d | RFX6 | hPSCs differentiated IPCs | human | [25] | |
| let-7e | RFX6 | hPSCs differentiated IPCs | human | [25] | |
| miR-21 | SOX6; RBJ | pancreatic progenitor cells | human | [26] | |
| miR-9 | - | human fetal islets | human | [27] | |
| miR-376 | - | human fetal islets | human | [27] | |
| Insulin secretion | miR-375 | Mtpn | MIN-6 cells | mouse | [28] |
| miR-184 | Ago 2 | MIN-6 cells | mouse | [29] | |
| miR-7a | - | β-cells of diabetic mice | mouse | [30] | |
| miR-29a | Stx-1a; Mct1 | rat islets; mouse islets | rat; mouse | [31,32] | |
| miR-187 | HIPK3 | human islets from T2DM patients | human | [33] | |
| miR-30a | β2; NeuroD1 | rat islets and INS-1 cells | rat | [34] | |
| miR-124 | Rab27a; Foxa2 | human islets; MIN-6 cells | human; mouse | [35,36] | |
| miR-33 | Abca1 | mouse islets; MIN-6 cells | mouse | [37] |
| Cell Processes | miRNA | Putative Targets | Cells or Tissue Studied | Species | References |
|---|---|---|---|---|---|
| IR | miR-15b | INSR | hepatocytes of diabetic mice | mouse | [39] |
| miR-195 | HepG2 cells | human | [40] | ||
| miR-29a | IRS-1 | L6 cells | rat | [41] | |
| miR-7 | C2C12 cells | mouse | [42] | ||
| miR-96 | SK-Hep1 cells | human | [43] | ||
| miR-126 | adipose tissue | human | [44] | ||
| miR-135a | Irs-2 | C2C12 cells | mouse | [46] | |
| miR-378 | PI3K | hepatocytes of miR-378/378 * knockout mice | mouse | [47] | |
| miR-320 | 3T3-L1 cells | mouse | [48] | ||
| miR-199a | GLUT4 | L6 cells | rat | [52] | |
| miR-223 | human differentiated adipocytes | human | [53] | ||
| miR-93 | subcutaneous adipose tissue | human | [54] | ||
| miR-499 | PTEN | NCTC 1469 cells; livers of db/db mice | mouse | [49] | |
| miR-26b | human insulin-resistant viceral adipocytes | human | [50] | ||
| miR-301a | NCTC 1469 cells | mouse | [51] | ||
| miR-143 | Orp8 | HL-1 cells | mouse | [55] | |
| livers of obese mice | mouse | [56] | |||
| miR-181a | SIRT1 | HepG2 cells | human | [57] | |
| miR-543 | HepG2 cells | human | [57] | ||
| miR-29 | Pparδ | C2C12 cells | mouse | [53] | |
| miR-103/107 | Cav-1 | livers and adipose tissues of diet induced obesity mice | mouse | [45] | |
| miR-802 | Hnf1β | Hepa1-6 cells | mouse | [58] | |
| miR-106b | Mfn2 | C2C12 cells | mouse | [59] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, J.; Xing, W.; Xie, L. Regulatory Roles of MicroRNAs in Diabetes. Int. J. Mol. Sci. 2016, 17, 1729. https://doi.org/10.3390/ijms17101729
Feng J, Xing W, Xie L. Regulatory Roles of MicroRNAs in Diabetes. International Journal of Molecular Sciences. 2016; 17(10):1729. https://doi.org/10.3390/ijms17101729
Chicago/Turabian StyleFeng, Juan, Wanli Xing, and Lan Xie. 2016. "Regulatory Roles of MicroRNAs in Diabetes" International Journal of Molecular Sciences 17, no. 10: 1729. https://doi.org/10.3390/ijms17101729






