Role of miR-34a-5p in Hematopoietic Progenitor Cells Proliferation and Fate Decision: Novel Insights into the Pathogenesis of Primary Myelofibrosis
Abstract
:1. Introduction
2. Results
2.1. miR-34a-5p Expression Levels in CD34+ Cells from PMF Patients and HDs
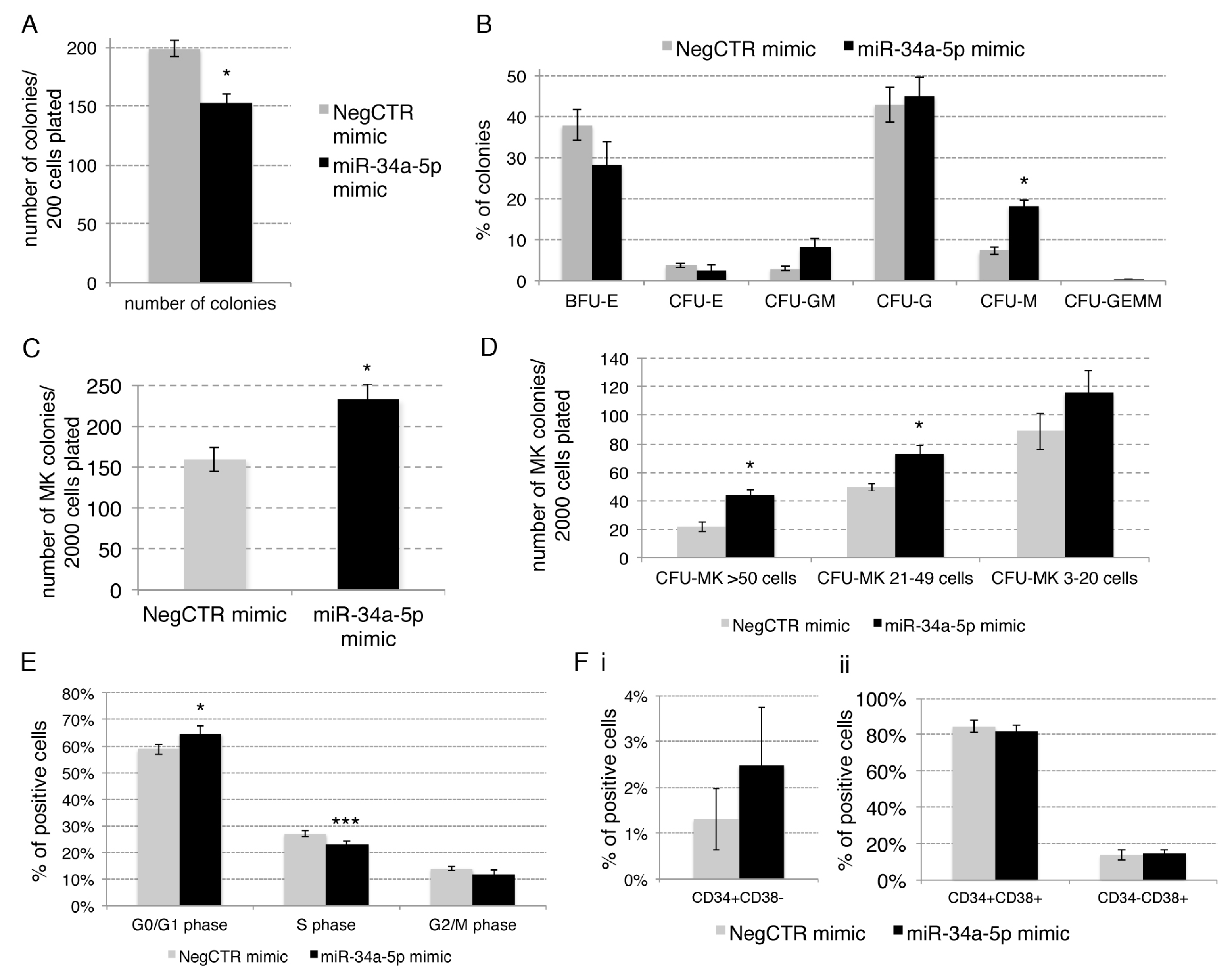
2.2. Effects of miR-34a-5p Overexpression on HPCs Proliferation and Clonogenic Efficiency
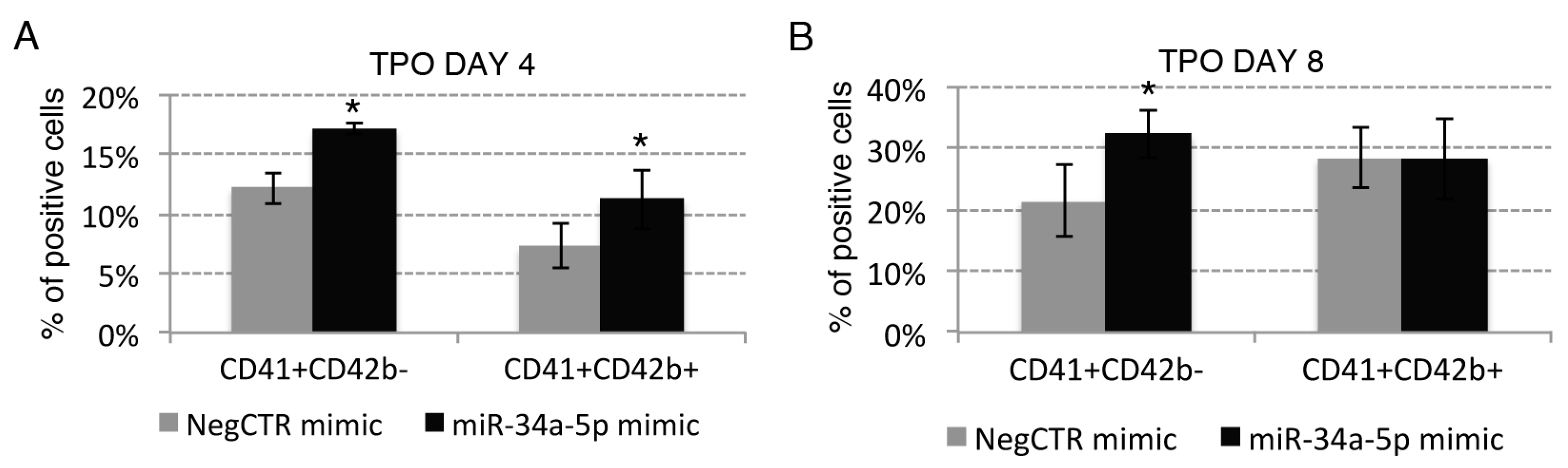
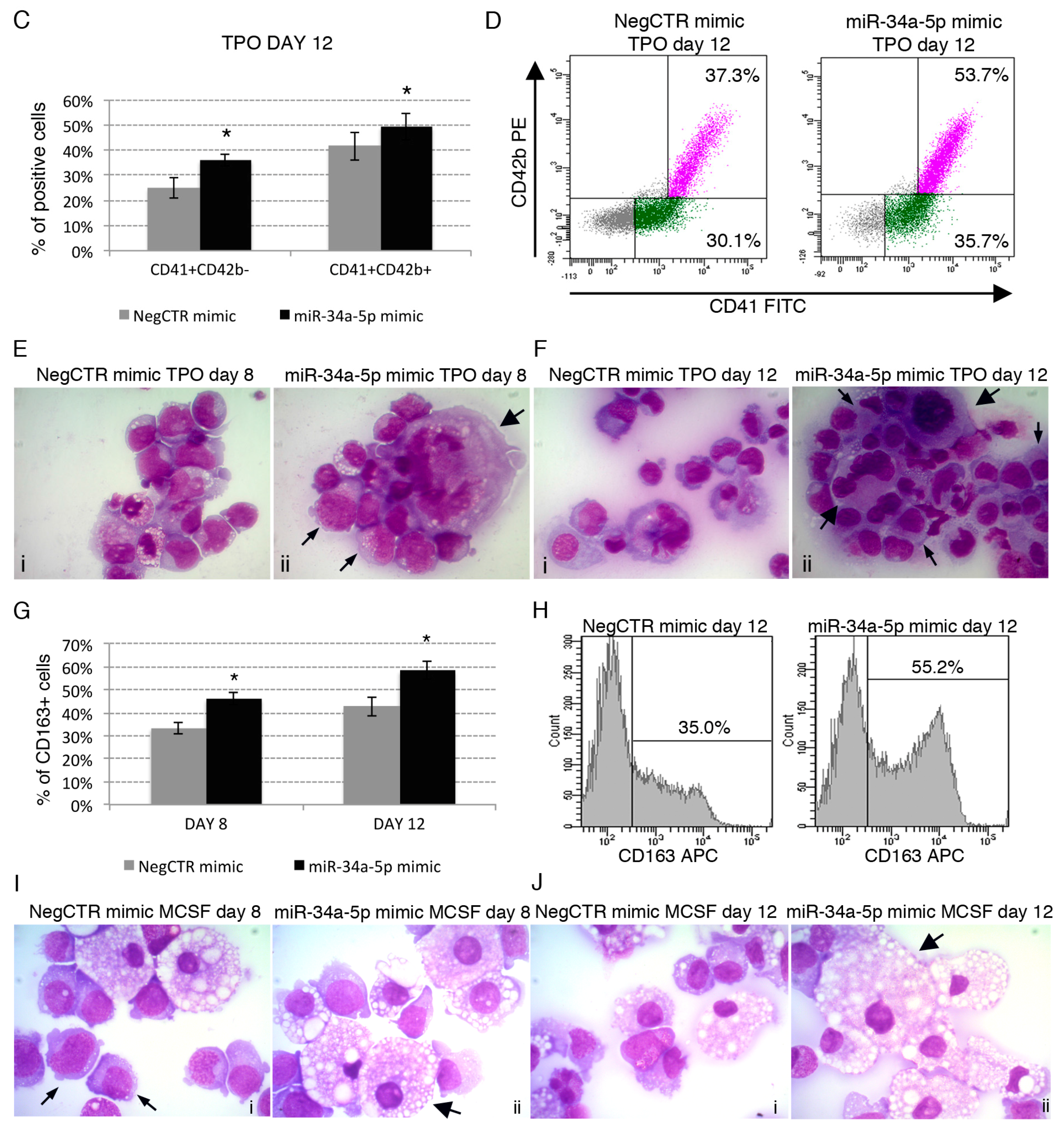
2.3. Effects of miR-34a-5p Overexpression on HPCs Lineage Choice
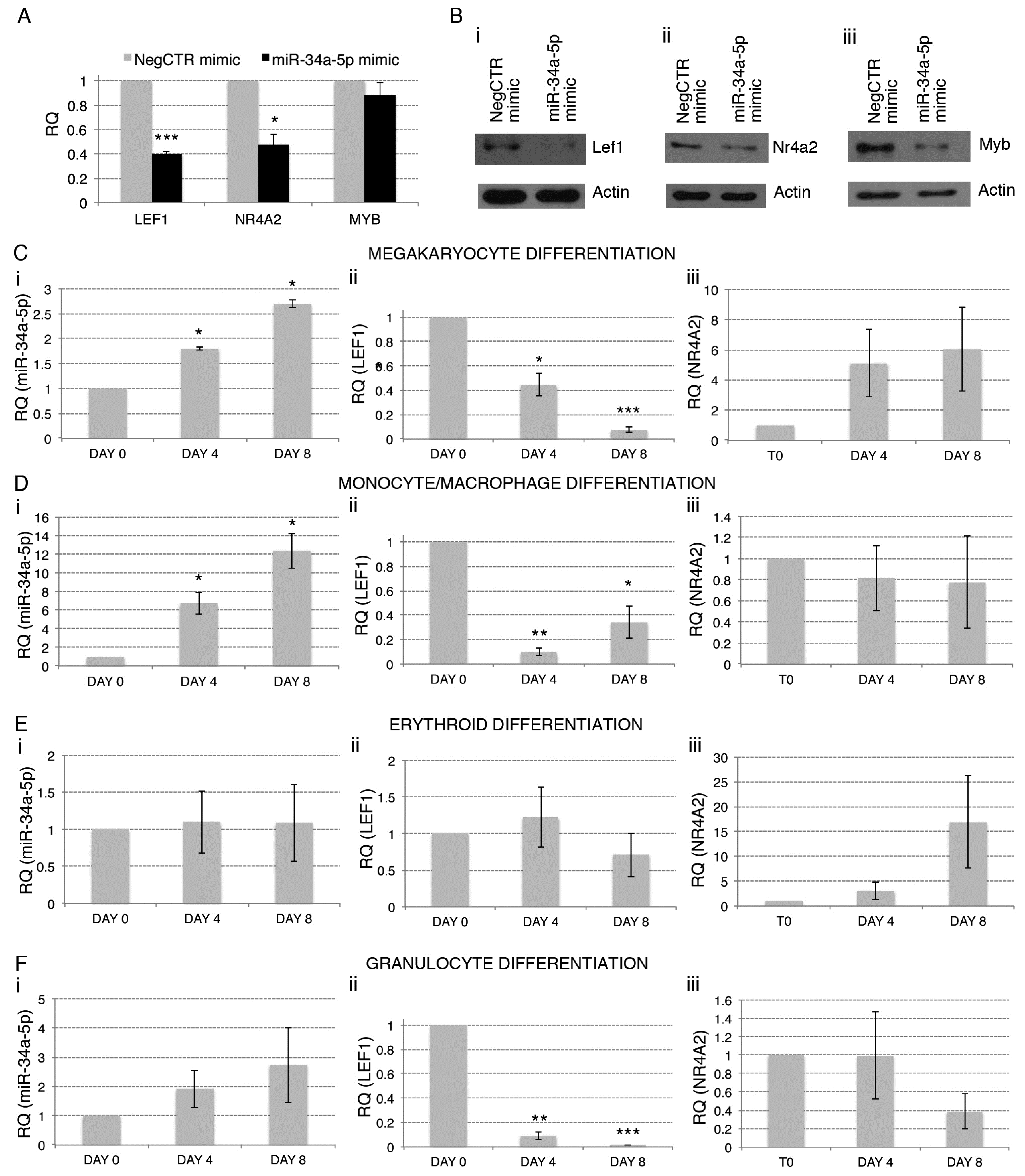
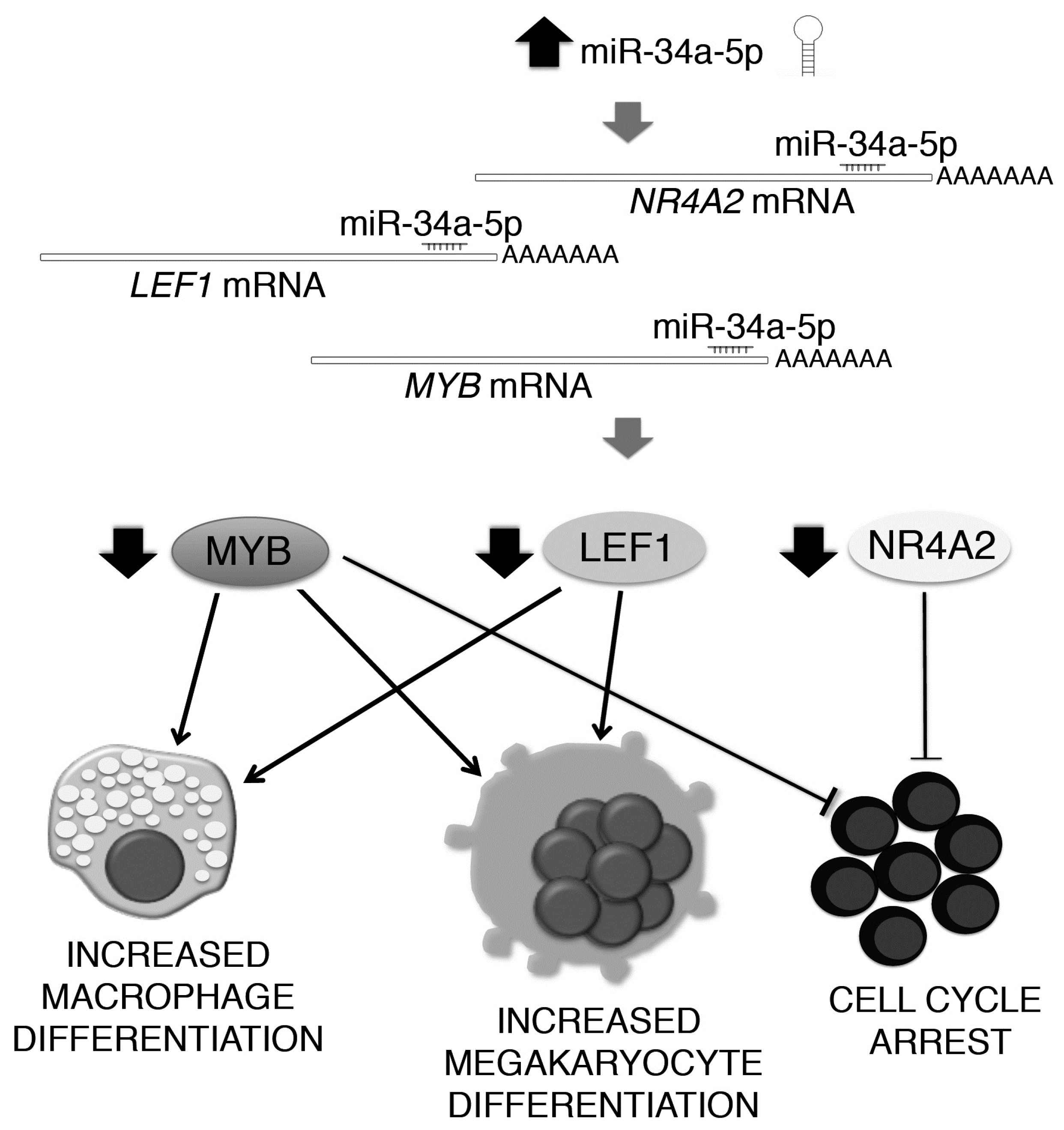
2.4. Gene Expression Profile (GEP) of miR-34a-5p-Overexpressing Cells and Identification of miR-34a-5p Targets
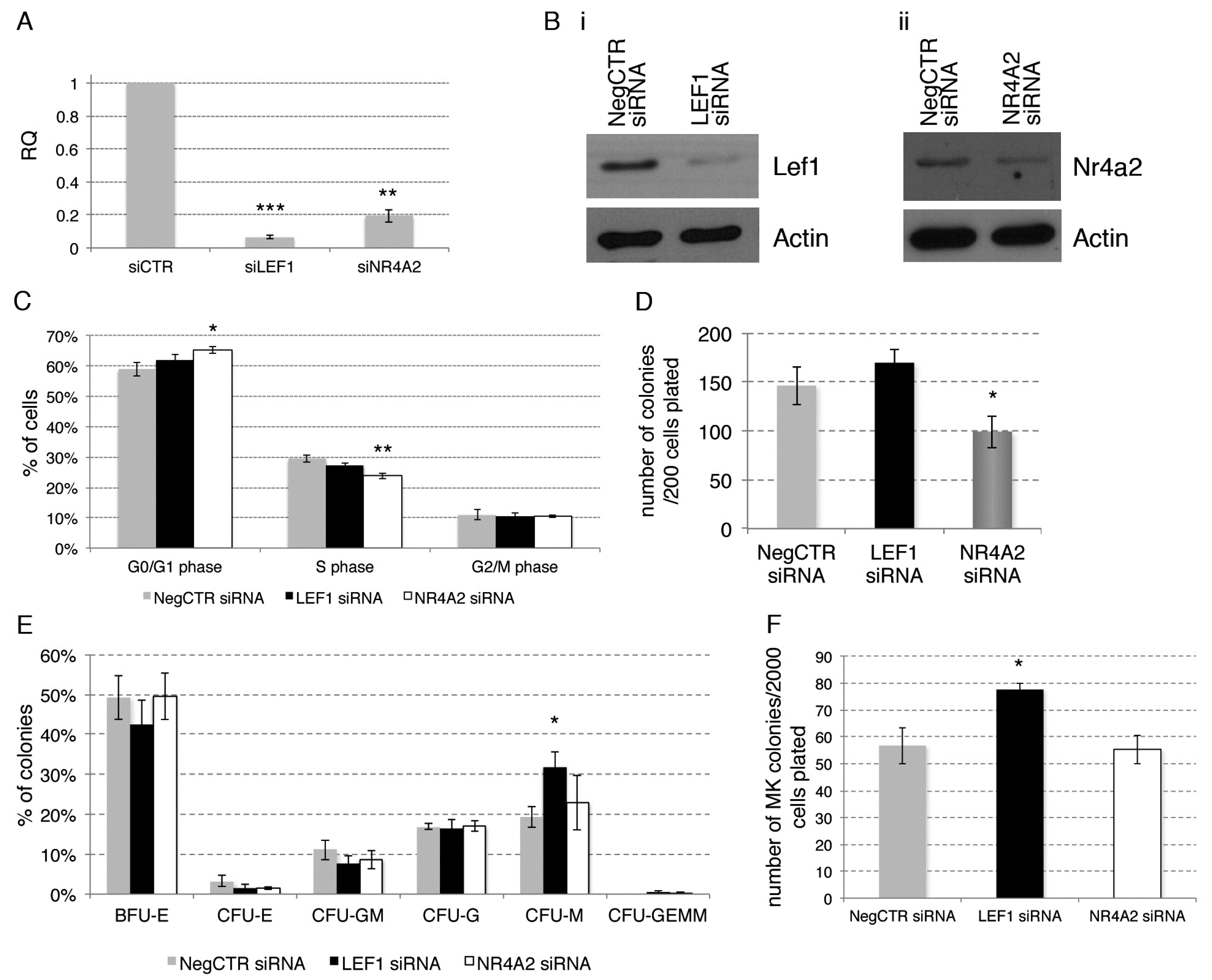
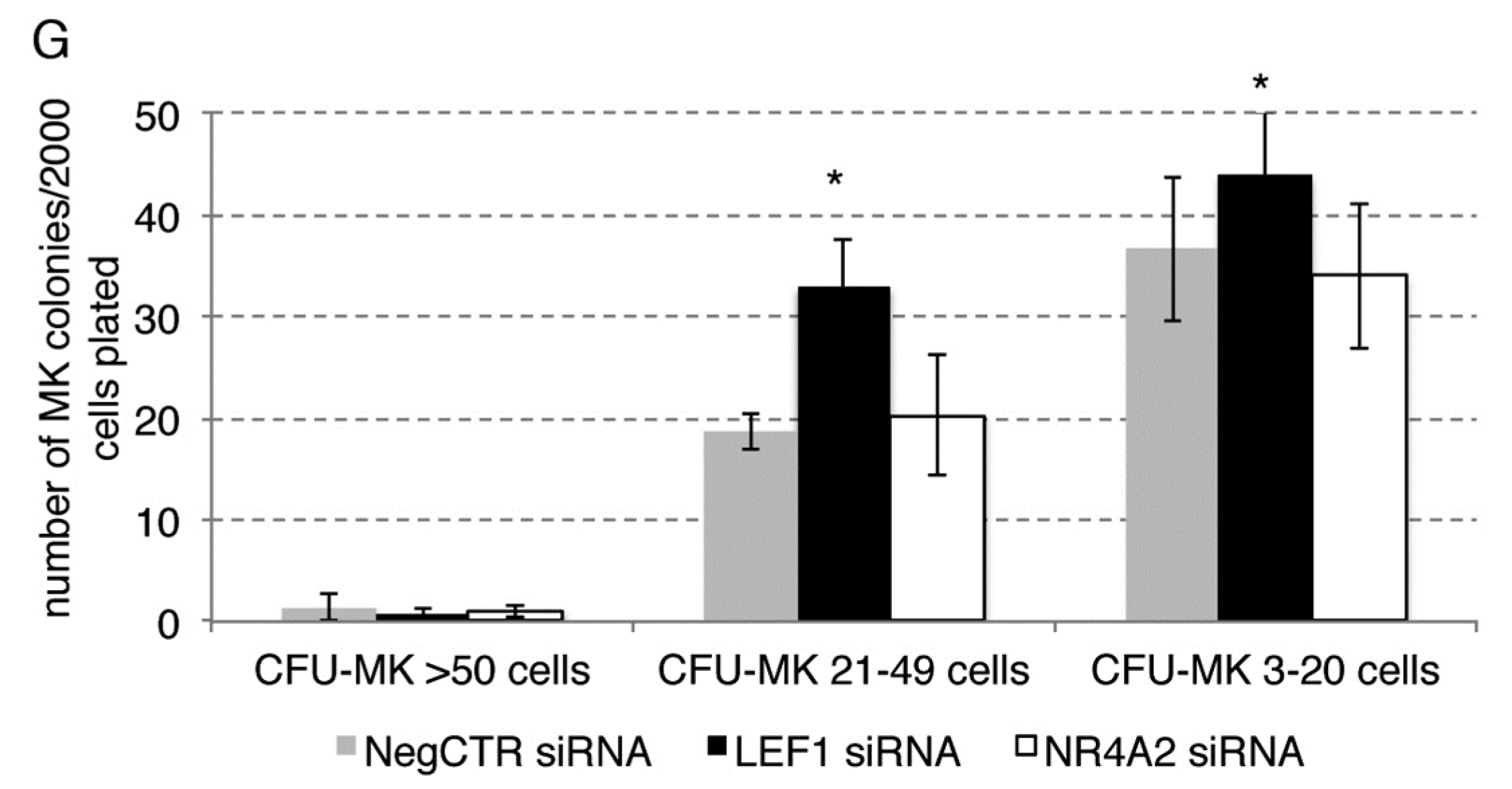
2.5. Effects of LEF1 and NR4A2 Silencing on HPCs Proliferation and Clonogenic Efficiency
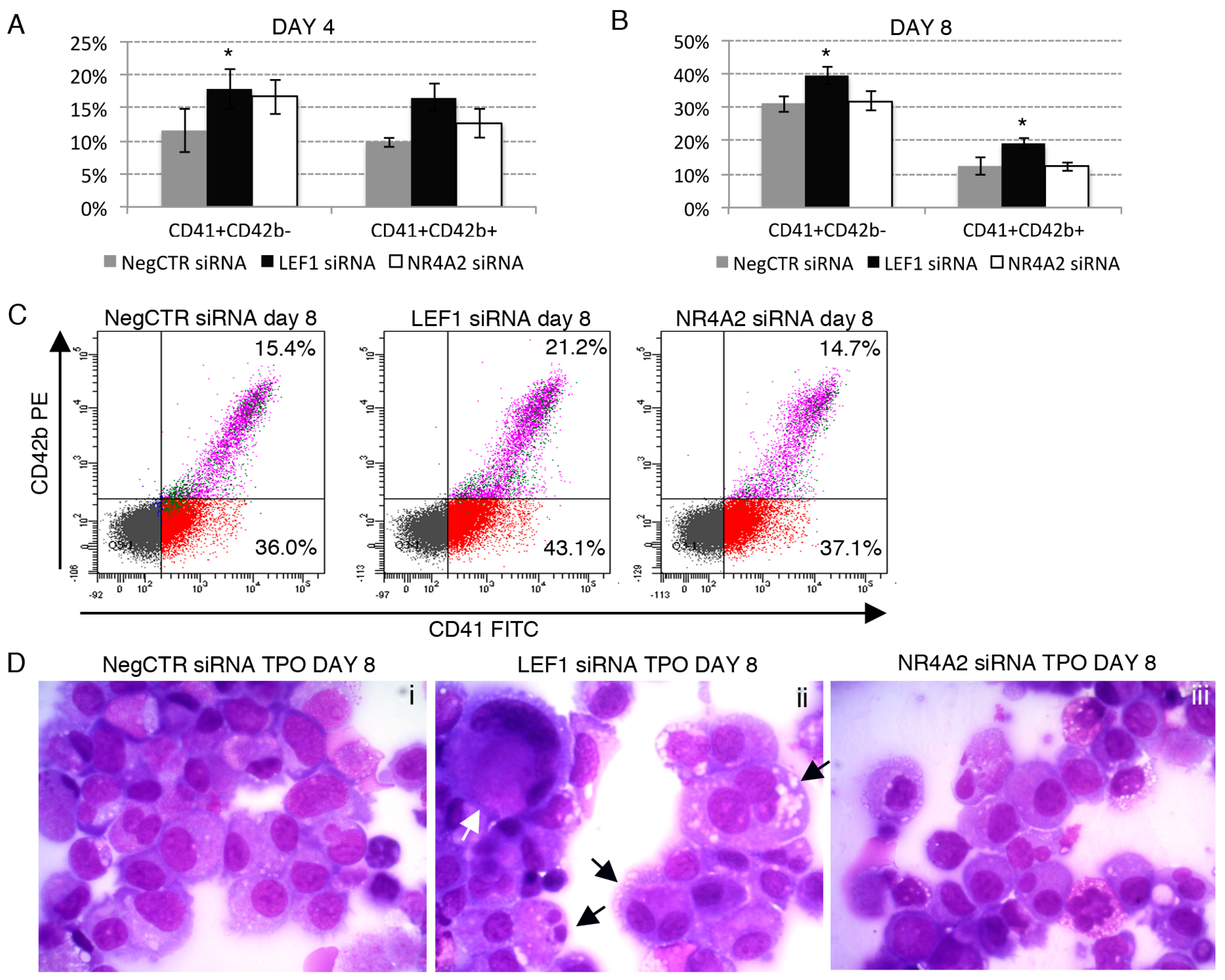
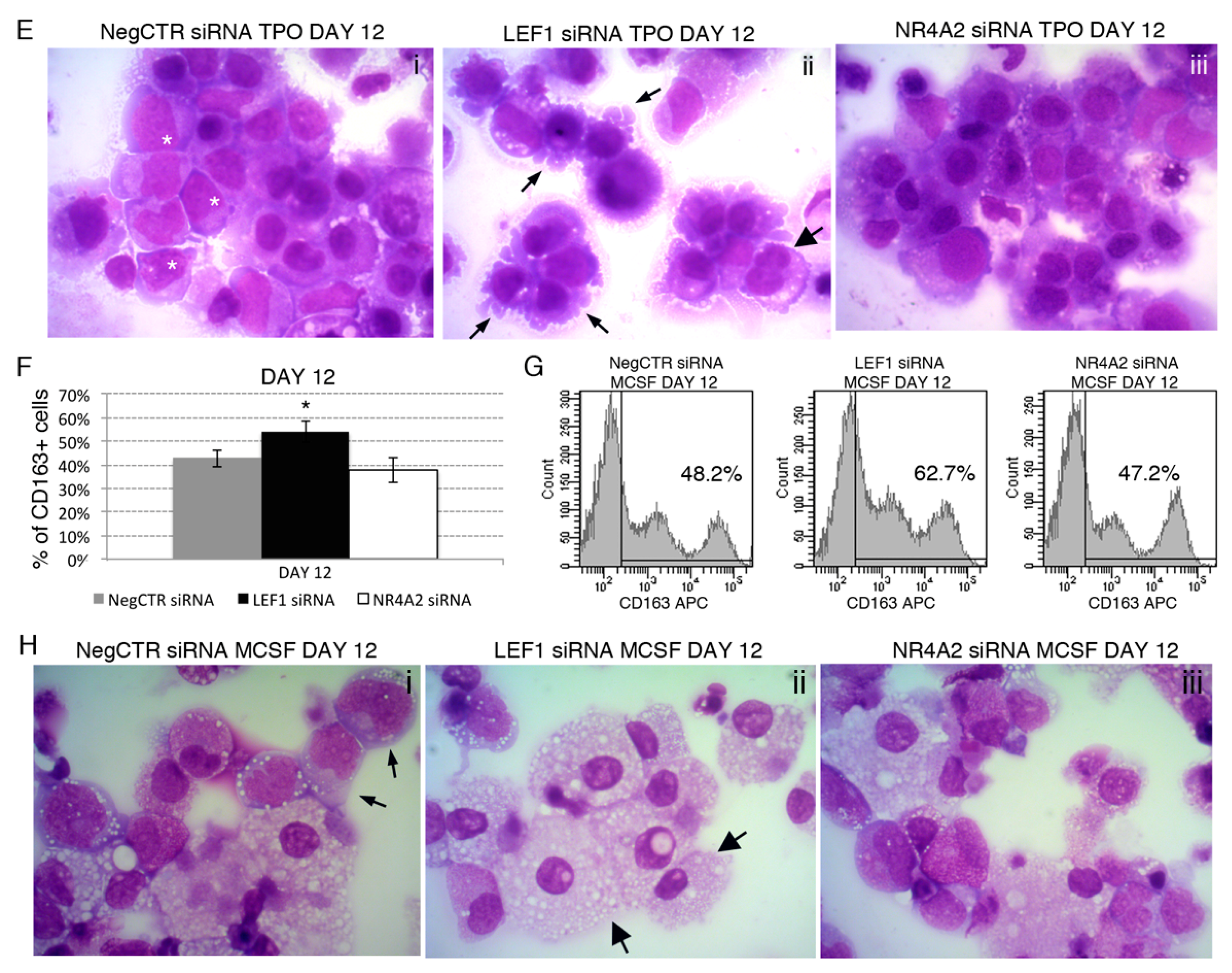
2.6. Effects of LEF1 and NR4A2 on the HPCs Lineage Choice
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. miR-34a-5p Expression Analysis in PMF and HD CD34+ Cells
4.3. Human CD34+ Hematopoietic Progenitor Cells (HPCs) Purification
4.4. miR-34a-5p, LEF1 and NR4A2 Kinetics during the Erythroid, Megakaryocyte, Granulocyte and Mono-Macrophage Differentiation of CD34+ HPCs
4.5. Nucleofection of CD34+ Cells
4.6. Methylcellulose- and Collagen-Based Clonogenic Assays
4.7. Cell Cycle Analysis
4.8. Morphological and Immunophenotypic Analysis
4.9. RNA Extraction
4.10. Gene Expression Profiling (GEP)
4.11. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
4.12. Western Blot
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, P.; Takizawa, H.; Kubovcakova, L.; Guo, G.; Hao-Shen, H.; Dirnhofer, S.; Orkin, S.H.; Manz, M.G.; Skoda, R.C. Myeloproliferative neoplasms can be initiated from a single hematopoietic stem cell expressing JAK2-V617F. J. Exp. Med. 2014, 211, 2213–2230. [Google Scholar] [CrossRef] [PubMed]
- Barbui, T.; Thiele, J.; Gisslinger, H.; Finazzi, G.; Vannucchi, A.M.; Tefferi, A. The 2016 revision of WHO classification of myeloproliferative neoplasms: Clinical and molecular advances. Blood Rev. 2016, 30, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Desterke, C.; Martinaud, C.; Ruzehaji, N.; Le Bousse-Kerdiles, M.C. Inflammation as a Keystone of Bone Marrow Stroma Alterations in Primary Myelofibrosis. Mediat. Inflamm. 2015, 2015, 415024. [Google Scholar] [CrossRef] [PubMed]
- Le Bousse-Kerdiles, M.C. Primary myelofibrosis and the “bad seeds in bad soil“ concept. Fibrogenes. Tissue Repair. 2012, 5, S20. [Google Scholar]
- Rameshwar, P.; Denny, T.N.; Stein, D.; Gascon, P. Monocyte adhesion in patients with bone marrow fibrosis is required for the production of fibrogenic cytokines. Potential role for interleukin-1 and TGF-β. J. Immunol. 1994, 153, 2819–2830. [Google Scholar] [PubMed]
- Rameshwar, P.; Narayanan, R.; Qian, J.; Denny, T.N.; Colon, C.; Gascon, P. NF-κB as a central mediator in the induction of TGF-β in monocytes from patients with idiopathic myelofibrosis: An inflammatory response beyond the realm of homeostasis. J. Immunol. 2000, 165, 2271–2277. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Norfo, R.; Pennucci, V.; Zini, R.; Manfredini, R. Genomic landscape of megakaryopoiesis and platelet function defects. Blood 2016, 127, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Ortmann, C.A.; Kent, D.G.; Nangalia, J.; Silber, Y.; Wedge, D.C.; Grinfeld, J.; Baxter, E.J.; Massie, C.E.; Papaemmanuil, E.; Menon, S.; et al. Effect of mutation order on myeloproliferative neoplasms. N. Engl. J. Med. 2015, 372, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, P.; Karow, A.; Nienhold, R.; Looser, R.; Hao-Shen, H.; Nissen, I.; Girsberger, S.; Lehmann, T.; Passweg, J.; Stern, M.; et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood 2014, 123, 2220–2228. [Google Scholar] [CrossRef] [PubMed]
- Tenedini, E.; Bernardis, I.; Artusi, V.; Artuso, L.; Roncaglia, E.; Guglielmelli, P.; Pieri, L.; Bogani, C.; Biamonte, F.; Rotunno, G.; et al. Targeted cancer exome sequencing reveals recurrent mutations in myeloproliferative neoplasms. Leukemia 2014, 28, 1052–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norfo, R.; Zini, R.; Pennucci, V.; Bianchi, E.; Salati, S.; Guglielmelli, P.; Bogani, C.; Fanelli, T.; Mannarelli, C.; Rosti, V.; et al. miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: Role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood 2014, 124, e21–e32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, F.; Gutman, D.; Meire, E.; Caceres, M.; Rigoutsos, I.; Bentwich, Z.; Lieberman, J. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood 2009, 114, 2181–2192. [Google Scholar] [CrossRef] [PubMed]
- Von Laer, D.; Corovic, A.; Vogt, B.; Fehse, B.; Roscher, S.; Rimek, A.; Baum, C.; Ostertag, W. Loss of CD38 antigen on CD34+CD38+ cells during short-term culture. Leukemia 2000, 14, 947–948. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, Y.; Daniels, G.; Sfanos, K.; de Marzo, A.; Wei, J.; Li, X.; Chen, W.; Wang, J.; Zhong, X.; et al. LEF1 Targeting EMT in Prostate Cancer Invasion Is Regulated by miR-34a. Mol. Cancer Res. 2015, 13, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Beard, J.A.; Tenga, A.; Hills, J.; Hoyer, J.D.; Cherian, M.T.; Wang, Y.D.; Chen, T. The orphan nuclear receptor NR4A2 is part of a p53-microRNA-34 network. Sci. Rep. 2016, 6, 25108. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Zini, R.; Salati, S.; Tenedini, E.; Norfo, R.; Tagliafico, E.; Manfredini, R.; Ferrari, S. c-Myb supports erythropoiesis through the transactivation of KLF1 and LMO2 expression. Blood 2010, 116, e99–e110. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Bulgarelli, J.; Ruberti, S.; Rontauroli, S.; Sacchi, G.; Norfo, R.; Pennucci, V.; Zini, R.; Salati, S.; Prudente, Z.; et al. MYB controls erythroid vs. megakaryocyte lineage fate decision through the miR-486–3p-mediated downregulation of MAF. Cell Death Differ. 2015, 22, 1906–1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, A.; Ruike, Y.; Terasawa, K.; Shimizu, K.; Tsujimoto, G. MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during megakaryocytic differentiation of K562 cells. Mol. Pharmacol. 2010, 77, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Desterke, C.; Bilhou-Nabera, C.; Guerton, B.; Martinaud, C.; Tonetti, C.; Clay, D.; Guglielmelli, P.; Vannucchi, A.; Bordessoule, D.; Hasselbalch, H.; et al. FLT3-mediated p38-MAPK activation participates in the control of megakaryopoiesis in primary myelofibrosis. Cancer Res. 2011, 71, 2901–2915. [Google Scholar] [CrossRef] [PubMed]
- Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell 1999, 97, 161–163. [Google Scholar]
- Philips, A.; Lesage, S.; Gingras, R.; Maira, M.H.; Gauthier, Y.; Hugo, P.; Drouin, J. Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol. Cell. Biol. 1997, 17, 5946–5951. [Google Scholar] [CrossRef] [PubMed]
- Philips, A.; Maira, M.; Mullick, A.; Chamberland, M.; Lesage, S.; Hugo, P.; Drouin, J. Antagonism between Nur77 and glucocorticoid receptor for control of transcription. Mol. Cell. Biol. 1997, 17, 5952–5959. [Google Scholar] [CrossRef] [PubMed]
- Sirin, O.; Lukov, G.L.; Mao, R.; Conneely, O.M.; Goodell, M.A. The orphan nuclear receptor Nurr1 restricts the proliferation of haematopoietic stem cells. Nat. Cell Biol. 2010, 12, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Li, W.; Sheng, Y.; Li, L.; Huang, Y.; Zhang, Z.; Zhu, T.; Peace, D.; Quigley, J.G.; Wu, W.; et al. The transcription factor Foxm1 is essential for the quiescence and maintenance of hematopoietic stem cells. Nat. Immunol. 2015, 16, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Llopis, S.; Singleton, B.; Duplessis, T.; Carrier, L.; Rowan, B.; Williams, C. Dichotomous roles for the orphan nuclear receptor NURR1 in breast cancer. BMC Cancer 2013, 13, 139. [Google Scholar] [CrossRef] [PubMed]
- Zu, G.; Yao, J.; Ji, A.; Ning, S.; Luo, F.; Li, Z.; Feng, D.; Rui, Y.; Li, Y.; Wang, G.; et al. Nurr1 promotes intestinal regeneration after ischemia/reperfusion injury by inhibiting the expression of p21 (Waf1/Cip1). J. Mol. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Chachoua, I.; Pecquet, C.; El-Khoury, M.; Nivarthi, H.; Albu, R.I.; Marty, C.; Gryshkova, V.; Defour, J.P.; Vertenoeil, G.; Ngo, A.; et al. Thrombopoietin receptor activation by myeloproliferative neoplasm associated calreticulin mutants. Blood 2016, 127, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Marty, C.; Pecquet, C.; Nivarthi, H.; El-Khoury, M.; Chachoua, I.; Tulliez, M.; Villeval, J.L.; Raslova, H.; Kralovics, R.; Constantinescu, S.N.; et al. Calreticulin mutants in mice induce an MPL-dependent thrombocytosis with frequent progression to myelofibrosis. Blood 2016, 127, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Eastman, Q.; Grosschedl, R. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr. Opin. Cell Biol. 1999, 11, 233–240. [Google Scholar] [CrossRef]
- Macaulay, I.C.; Thon, J.N.; Tijssen, M.R.; Steele, B.M.; MacDonald, B.T.; Meade, G.; Burns, P.; Rendon, A.; Salunkhe, V.; Murphy, R.P.; et al. Canonical Wnt signaling in megakaryocytes regulates proplatelet formation. Blood 2013, 121, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Ubreva, J.; Ciudad, L.; van Oevelen, C.; Parra, M.; Graf, T.; Ballestar, E. C/EBPa-mediated activation of microRNAs 34a and 223 inhibits Lef1 expression to achieve efficient reprogramming into macrophages. Mol. Cell. Biol. 2014, 34, 1145–1157. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.J.; Yang, Q.; Goldenson, B.; Malinge, S.; Lasho, T.; Schneider, R.K.; Breyfogle, L.J.; Schultz, R.; Gilles, L.; Koppikar, P.; et al. Targeting megakaryocytic-induced fibrosis in myeloproliferative neoplasms by AURKA inhibition. Nat. Med. 2015, 21, 1473–1480. [Google Scholar] [PubMed]
- Wagner-Ballon, O.; Pisani, D.F.; Gastinne, T.; Tulliez, M.; Chaligne, R.; Lacout, C.; Aurade, F.; Villeval, J.L.; Gonin, P.; Vainchenker, W.; et al. Proteasome inhibitor bortezomib impairs both myelofibrosis and osteosclerosis induced by high thrombopoietin levels in mice. Blood 2007, 110, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Bonta, P.I.; van Tiel, C.M.; Vos, M.; Pols, T.W.; van Thienen, J.V.; Ferreira, V.; Arkenbout, E.K.; Seppen, J.; Spek, C.A.; van der Poll, T.; et al. Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 2288–2294. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, J.; Huo, Y.; Lu, J.; Wan, L.; Yang, Q.; Huang, J.; Gan, R.; Guo, C. Adenovirus-mediated expression of orphan nuclear receptor NR4A2 targeting hepatic stellate cell attenuates liver fibrosis in rats. Sci. Rep. 2016, 6, 33593. [Google Scholar] [CrossRef] [PubMed]
- Rampal, R.; Al-Shahrour, F.; Abdel-Wahab, O.; Patel, J.P.; Brunel, J.P.; Mermel, C.H.; Bass, A.J.; Pretz, J.; Ahn, J.; Hricik, T.; et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood 2014, 123, e123–e133. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Tenedini, E.; Roncaglia, E.; Ferrari, F.; Orlandi, C.; Bianchi, E.; Bicciato, S.; Tagliafico, E.; Ferrari, S. Integrated analysis of microRNA and mRNA expression profiles in physiological myelopoiesis: Role of hsa-mir-299–5p in CD34+ progenitor cells commitment. Cell Death Dis. 2010, 1, e28. [Google Scholar] [CrossRef] [PubMed]
- Kandilci, A.; Grosveld, G.C. Reintroduction of CEBPA in MN1-overexpressing hematopoietic cells prevents their hyperproliferation and restores myeloid differentiation. Blood 2009, 114, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, I.; Migliorati, G.; Pagliacci, M.C.; Grignani, F.; Riccardi, C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods. 1991, 139, 271–279. [Google Scholar] [CrossRef]

| Probeset ID | Gene Symbol | Gene Title | RefSeq Transcript ID | FC miR-34a-5p Mimic vs. NegCTR Mimic CD34+ Cells | TargetScan-Predicted miR-34a-5p Target | Modulation in PMF vs. HD CD34+ Cells |
|---|---|---|---|---|---|---|
| 11734171_at | PRTN3 | proteinase 3 | NM_002777 | −3.81 | − | − |
| 11728026_x_at | IGLL1 | immunoglobulin lambda-like polypeptide 1 | NM_020070; NM_152855 | −3.22 | − | − |
| 11756600_a_at | TPD52 | tumor protein D52 | NM_001025252;NM_001025253; NM_005079 | −2.93 | + | − |
| 11729643_s_at | TPD52 | tumor protein D52 | NM_001025252; NM_001025253; NM_005079 | −2.89 | + | − |
| 11754659_x_at | TPD52 | tumor protein D52 | NM_001025252;NM_001025253;NM_005079 | −2.69 | + | − |
| 11727965_at | ELANE | elastase, neutrophil expressed | NM_001972 | −2.65 | − | − |
| 11715306_s_at | AREG; AREGB | amphiregulin /// amphiregulin B | NM_001657; XM_001125684 | −2.61 | + | D |
| 11745205_s_at | TPD52 | tumor protein D52 | NM_001025252;NM_001025253; NM_005079 | −2.59 | + | − |
| 11723339_at | CTSG | cathepsin G | NM_001911 | −2.29 | − | − |
| 11726333_s_at | LEF1 | lymphoid enhancer-binding factor 1 | NM_001130713;NM_001130714;NM_001166119; NM_016269 | −2.29 | + | D |
| 11718477_a_at | STAR | steroidogenic acute regulatory protein | NM_000349; NM_001007243 | −2.24 | − | − |
| 11720051_at | SPOCK1 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 | NM_004598 | −2.17 | − | − |
| 11718479_x_at | STAR | steroidogenic acute regulatory protein | NM_000349; NM_001007243 | −2.17 | − | − |
| 11715245_s_at | IGLL1 | immunoglobulin lambda-like polypeptide 1 | NM_020070; NM_152855 | −2.15 | − | − |
| 11729641_a_at | TPD52 | tumor protein D52 | NM_001025252; NM_001025253; NM_005079 | −2.06 | + | − |
| 11725632_at | NR4A2 | nuclear receptor subfamily 4, group A, member 2 | NM_006186; NM_173171; NM_173172; NM_173173 | −2.03 | + | D |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianchi, E.; Ruberti, S.; Rontauroli, S.; Guglielmelli, P.; Salati, S.; Rossi, C.; Zini, R.; Tagliafico, E.; Vannucchi, A.M.; Manfredini, R. Role of miR-34a-5p in Hematopoietic Progenitor Cells Proliferation and Fate Decision: Novel Insights into the Pathogenesis of Primary Myelofibrosis. Int. J. Mol. Sci. 2017, 18, 145. https://doi.org/10.3390/ijms18010145
Bianchi E, Ruberti S, Rontauroli S, Guglielmelli P, Salati S, Rossi C, Zini R, Tagliafico E, Vannucchi AM, Manfredini R. Role of miR-34a-5p in Hematopoietic Progenitor Cells Proliferation and Fate Decision: Novel Insights into the Pathogenesis of Primary Myelofibrosis. International Journal of Molecular Sciences. 2017; 18(1):145. https://doi.org/10.3390/ijms18010145
Chicago/Turabian StyleBianchi, Elisa, Samantha Ruberti, Sebastiano Rontauroli, Paola Guglielmelli, Simona Salati, Chiara Rossi, Roberta Zini, Enrico Tagliafico, Alessandro Maria Vannucchi, and Rossella Manfredini. 2017. "Role of miR-34a-5p in Hematopoietic Progenitor Cells Proliferation and Fate Decision: Novel Insights into the Pathogenesis of Primary Myelofibrosis" International Journal of Molecular Sciences 18, no. 1: 145. https://doi.org/10.3390/ijms18010145






