Overexpression of AmRosea1 Gene Confers Drought and Salt Tolerance in Rice
Abstract
:1. Introduction
2. Results
2.1. Generation of Transgenic Rice Plants by Overexpressing AmRosea1
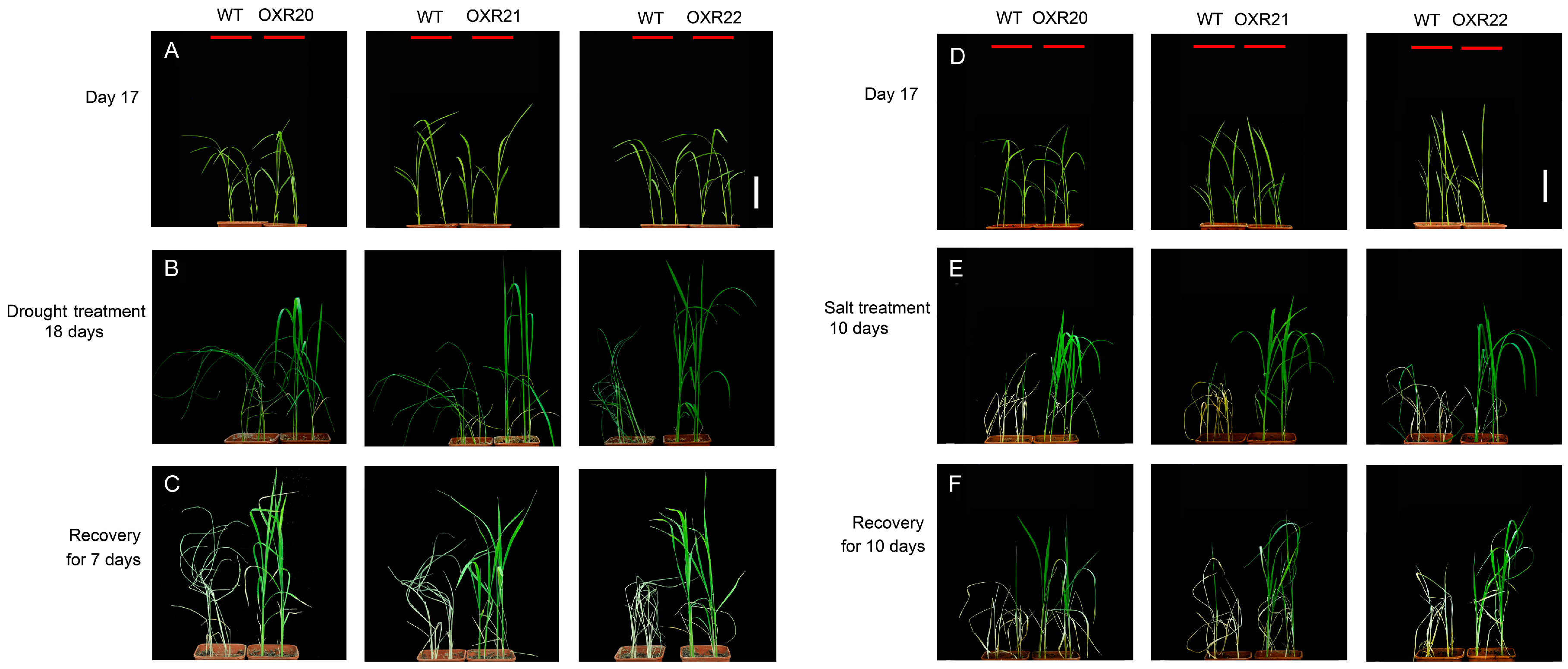
2.2. The Relationship between Overexpression of AmRosea1 and Improvement of Tolerance to Drought and Salt Stresses in Transgenic Rice
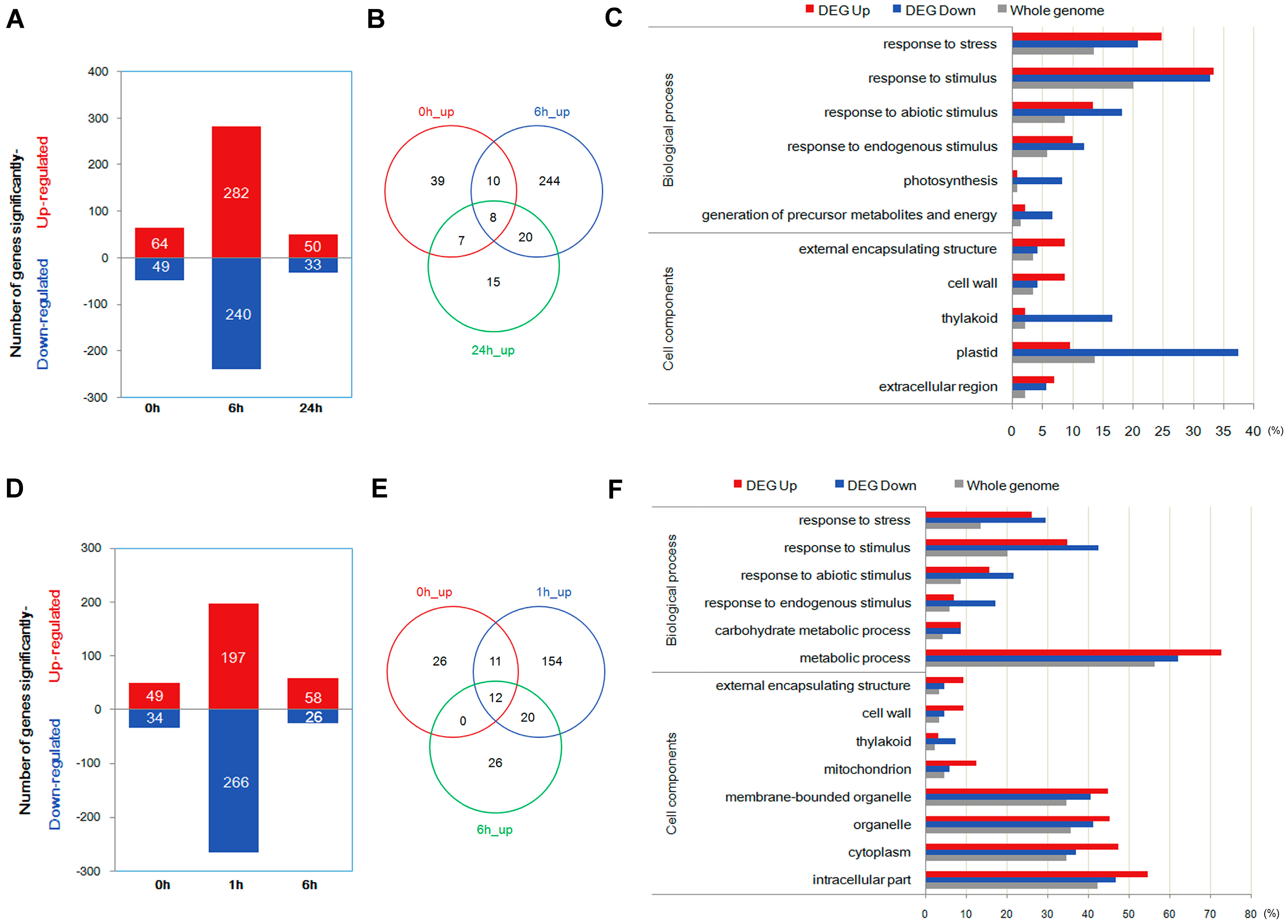
2.3. Quantification of Transcripts and Identification of Differentially Expressed Genes through Transcriptome Analysis
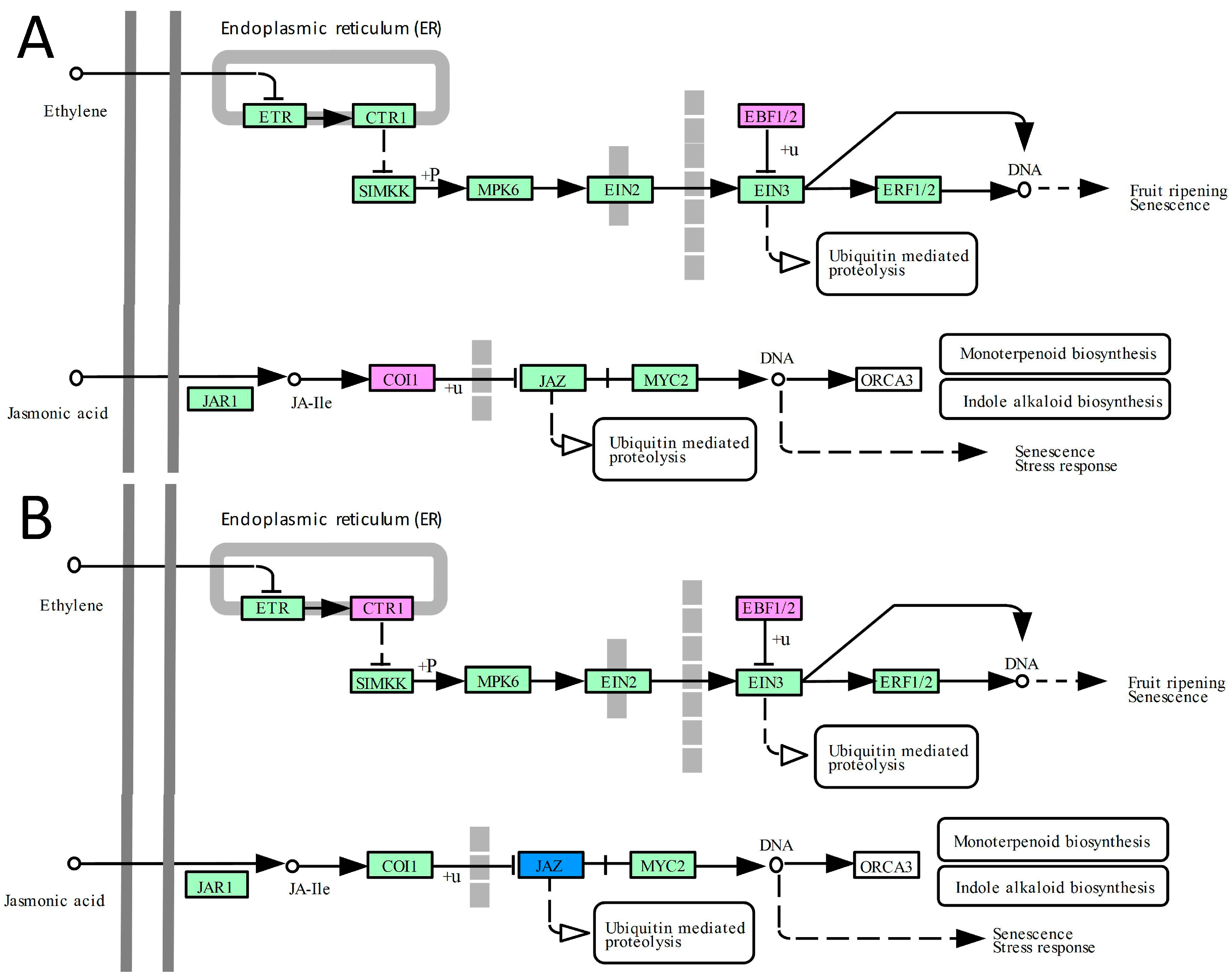
2.4. DEGs Related to Hormone Signal Transduction Pathways
2.5. DEGs Related to Protection from Osmotic Damage
2.6. DEGs Related to Protection from Oxidative Damage
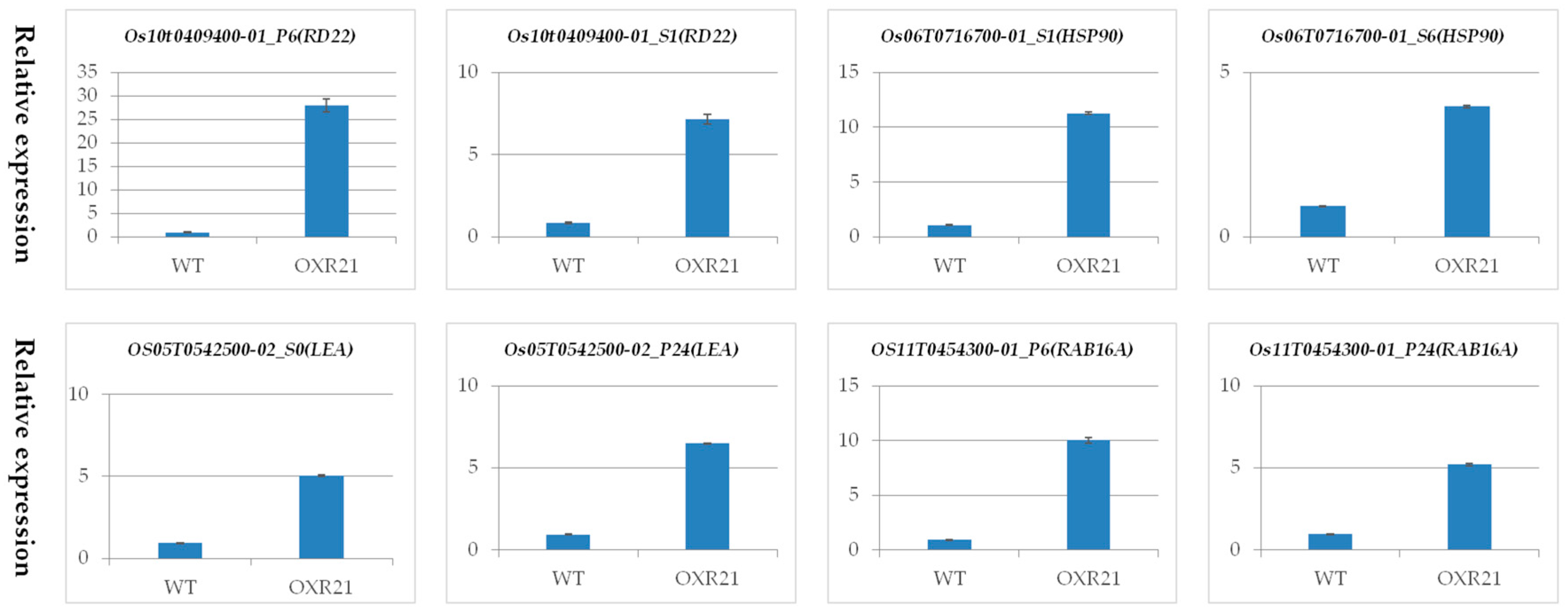
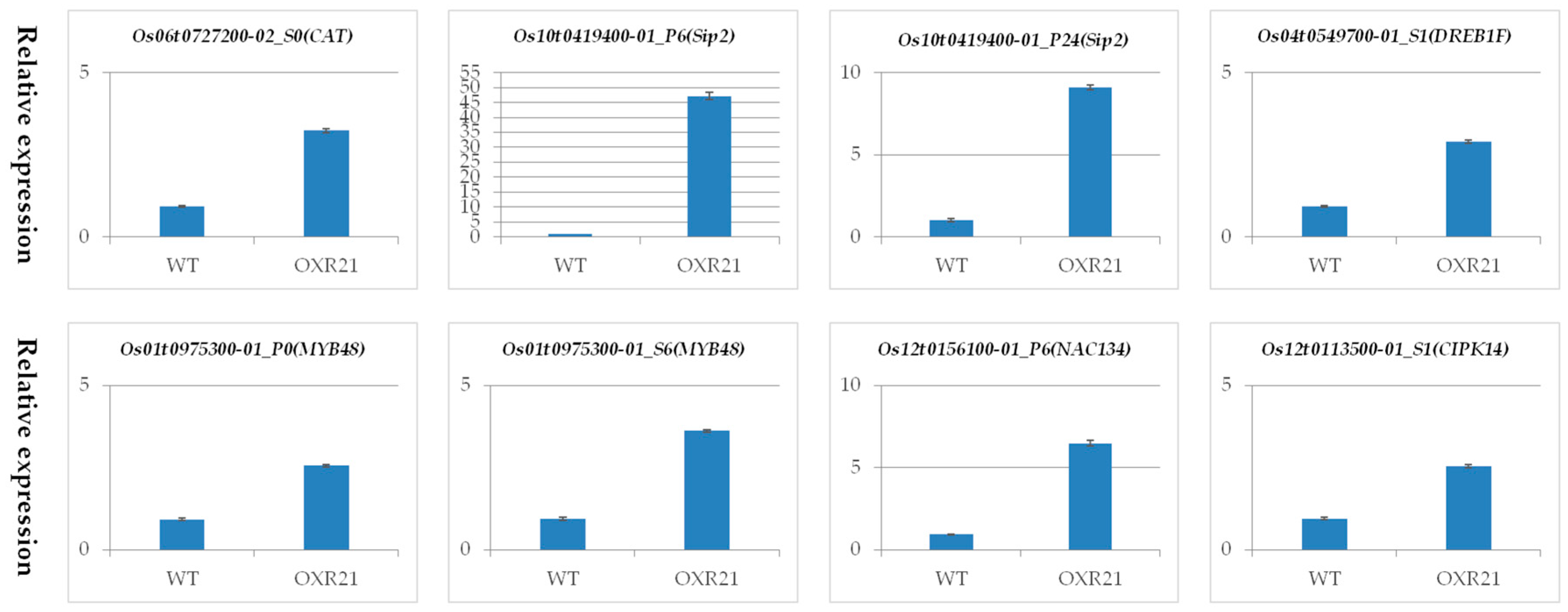
2.7. Verification of Differential Transcription Using Quantitative Real Time PCR (qRT-PCR)
3. Discussion
3.1. The Abiotic Tolerance of Transgenic Plants Extends AmROSEA1 Function beyond the Flavonoid Pathway
3.2. Some Unknown Function Genes and Several Abiotic Stress Response Genes Might Directly Relate to Abiotic Tolerance of Transgenic Plants
3.3. The Abiotic Tolerance of Transgenic Plants Is Gained from Regulations of Several Important Components of Plant Stress Defense Networks
3.4. The Ethylene and Jasmonate Pathways Contribute to the Improvement of Abiotic Tolerance in Transgenic Plants
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Plasmid Construction
4.3. Transformation of AmRosea1 and Screening Homozygote Lines
4.4. PCR Analysis
4.5. Anthocyanin and Total Flavonoid Content Quantification
4.6. Stress Tolerance Experiments
4.7. cDNA Library Construction and Illumina Sequencing
4.8. Analysis of RNA-Seq Data and Identification of DEGs
4.9. GO Enrichment and KEGG Mapping
4.10. qPCR Validation of Differential Transcription
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Feng, S.Q.; Wang, Y.L.; Yang, S.; Xu, Y.T.; Chen, X.S. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Lin-Wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamagishi, M.; Shimoyamada, Y.; Nakatsuka, T.; Masuda, K. Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of asiatic hybrid lily. Plant Cell Physiol. 2010, 51, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.S.; Xu, C.J.; Zhang, W.S.; Zhang, B.; Li, X.; Lin-Wang, K.; Ferguson, I.B.; Allan, A.C.; Chen, K.S. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 2010, 231, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.S.; Griffiths, H.M.; de Jong, D.M.; Cheng, S.; Bodis, M.; Kim, T.S.; de Jong, W.S. The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor. Appl. Genet. 2009, 120, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Kim, J.B.; Cho, K.J.; Cheon, C.I.; Sung, M.K.; Choung, M.G.; Roh, K.H. Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa). Plant Cell Rep. 2008, 27, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffe, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Griffiths, A.G.; Cousins, G.R.; Verry, I.M.; Williams, W.M. Anthocyanin leaf markings are regulated by a family of R2R3-MYB genes in the genus Trifolium. New Phytol. 2015, 205, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Fraser, L.G.; Seal, A.G.; Montefiori, M.; McGhie, T.K.; Tsang, G.K.; Datson, P.M.; Hilario, E.; Marsh, H.E.; Dunn, J.K.; Hellens, R.P.; et al. An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population. BMC Genom. 2013, 14, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowry, D.B.; Sheng, C.C.; Lasky, J.R.; Willis, J.H. Five anthocyanin polymorphisms are associated with an R2R3-MYB cluster in Mimulus guttatus (Phrymaceae). Am. J. Bot. 2012, 99, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Prescott, A.; Mackay, S.; Bartlett, J.; Vrijlandt, E. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1991, 1, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Peng, Q.; Zhao, J.; Owiti, A.; Ren, F.; Liao, L.; Wang, L.; Deng, X.; Jiang, Q.; Han, Y. Multiple R2R3-MYB transcription tactors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 2016, 7, 1557. [Google Scholar] [CrossRef] [PubMed]
- Schwinn, K.; Venail, J.; Shang, Y.J.; Mackay, S.; Alm, V.; Butelli, E.; Oyama, R.; Bailey, P.; Davies, K.; Martin, C. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 2006, 18, 831–851. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Li, S.; An, X.; Liu, X.; Qin, H.; Wang, D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genom. 2009, 36, 17–29. [Google Scholar] [CrossRef]
- Soltesz, A.; Vagujfalvi, A.; Rizza, F.; Kerepesi, I.; Galiba, G.; Cattivelli, L.; Coraggio, I.; Crosatti, C. The rice Osmyb4 gene enhances tolerance to frost and improves germination under unfavourable conditions in transgenic barley plants. J. Appl. Genet. 2012, 53, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.G.; Jang, C.S.; Kim, J.Y.; Kim, D.S.; Park, J.H.; Kim, D.Y.; Seo, Y.W. A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: Roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol. Plant. 2007, 129, 375–385. [Google Scholar] [CrossRef]
- Mao, X.; Jia, D.; Li, A.; Zhang, H.; Tian, S.; Zhang, X.; Jia, J.; Jing, R. Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Funct. Integr. Genom. 2011, 11, 445–465. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Wang, M.; Tian, Y.; He, W.; Han, L.; Xia, G. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 2012, 39, 7183–7192. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, W.; Lv, J.; Jia, Y.; Wang, M.; Xia, G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J. Exp. Bot. 2012, 63, 5873–5885. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Outchkourov, N.S.; Carollo, C.A.; Gomez-Roldan, V.; de Vos, R.C.H.; Bosch, D.; Hall, R.D.; Beekwilder, J. Control of anthocyanin and non-flavonoid compounds by anthocyanin-regulating MYB and bHLH transcription factors in Nicotiana benthamiana leaves. Front. Plant Sci. 2014, 5, 519. [Google Scholar] [CrossRef] [PubMed]
- Orzaez, D.; Medina, A.; Torre, S.; Patricia Fernandez-Moreno, J.; Luis Rambla, J.; Fernandez-del-Carmen, A.; Butelli, E.; Martin, C.; Granell, A. A visual reporter system for virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiol. 2009, 150, 1122–1134. [Google Scholar] [CrossRef] [PubMed]
- Schwinn, K.E.; Boase, M.R.; Bradley, J.M.; Lewis, D.H.; Deroles, S.C.; Martin, C.R.; Davies, K.M. MYB and bHLH transcription factor transgenes increase anthocyanin pigmentation in petunia and lisianthus plants, and the petunia phenotypes are strongly enhanced under field conditions. Front. Plant Sci. 2014, 5, 603. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Liu, C.; Zhang, Y.; Li, Y.; Yi, K.; Zhao, X.; Cui, M.L. The promoter structure differentiation of a MYB transcription factor RLC1 causes red leaf coloration in empire red leaf cotton under light. PLoS ONE 2013, 8, e77891. [Google Scholar] [CrossRef] [PubMed]
- Fridovich, I. Biological effects of the superoxide radical. Arch. Biochem. Biophys. 1986, 247, 1–11. [Google Scholar] [CrossRef]
- Imlay, J.A.; Linn, S. DNA damage and oxygen radical toxicity. Science 1988, 240, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Rus, A.M.; Bressan, R.A.; Hasegawa, P.M. Unraveling salt tolerance in crops. Nat. Genet. 2005, 37, 1029–1030. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 2014, 9, e92913. [Google Scholar] [CrossRef] [PubMed]
- Matus, J.T.; Aquea, F.; Espinoza, C.; Vega, A.; Cavallini, E.; dal Santo, S.; Cañón, P.; Rodríguez-Hoces de la Guardia, A.; Serrano, J.; Tornielli, G.B.; et al. Inspection of the grapevine BURP superfamily highlights an expansion of RD22 genes with distinctive expression features in berry development and ABA-mediated stress responses. PLoS ONE 2014, 9, e110372. [Google Scholar] [CrossRef] [PubMed]
- Harshavardhan, V.T.; van Son, L.; Seiler, C.; Junker, A.; Weigelt-Fischer, K.; Klukas, C.; Altmann, T.; Sreenivasulu, N.; Bäumlein, H.; Kuhlmann, M. AtRD22 and AtUSPL1, members of the plant-specific BURP domain family involved in Arabidopsis thaliana drought tolerance. PLoS ONE 2014, 9, e110065. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Morishita, H.; Urano, K.; Shiozaki, N.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Yoshiba, Y. Effects of free proline accumulation in petunias under drought stress. J. Exp. Bot. 2005, 56, 1975–1981. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.M.; Lin, Y.H. Peroxidase activity in relation to ethylene-induced rice (Oryza sativa L.) coleoptile elongation. Bot. Bull. Acad. Sin. 1996, 37, 239–245. [Google Scholar]
- Ismail, A.M.; Ella, E.S.; Vergara, G.V.; Mackill, D.J. Mechanisms associated with tolerance to flooding during germination and early seedling growth in rice (Oryza sativa). Ann. Bot. 2009, 103, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, S.K.; Pandey, A.; Pandey, G.K. The CBL-CIPK signaling module in plants: A mechanistic perspective. Physiol. Plant. 2015, 155, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Li, X.; Guo, M.; Deng, K.; Lin, J.; Tang, D.; Guo, X.; Liu, X. Regulation of salt and ABA responses by CIPK14, a calcium sensor interacting protein kinase in Arabidopsis. Sci. China Life Sci. 2008, 51, 391–401. [Google Scholar] [CrossRef]
- Deng, X.; Zhou, S.; Hu, W.; Feng, J.; Zhang, F.; Chen, L.; Huang, C.; Luo, Q.; He, Y.; Yang, G.; et al. Ectopic expression of wheat TaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiol. Plant. 2013, 149, 367–377. [Google Scholar] [PubMed]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.P.; Nakashima, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Chapter six—Plant gene networks in osmoticstress response: From genes to regulatory networks. Methods Enzymol. 2007, 428, 109–128. [Google Scholar] [PubMed]
- Wang, Q.; Guan, Y.; Wu, Y.; Chen, H.; Chen, F.; Chu, C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 2008, 67, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, D.; Li, H.; Wang, Y.; Zhang, Y.; Wood, A.J. EsDREB2B, a novel truncated DREB2-type transcription factor in the desert legume Eremosparton songoricum, enhances tolerance to multiple abiotic stresses in yeast and transgenic tobacco. BMC Plant Biol. 2014, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Lu, X.; Ma, B.; Chen, S.Y.; Zhang, J.S. Ethylene signaling in rice and Arabidopsis: Conserved and diverged aspects. Mol. Plant 2015, 8, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Smith, J.A.; Harberd, N.P.; Jiang, C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016, 91, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Lei, G.; Shen, M.; Li, Z.G.; Zhang, B.; Duan, K.X.; Wang, N.; Cao, Y.R.; Zhang, W.K.; Ma, B.; Ling, H.Q.; et al. EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ. 2011, 34, 1678–1692. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, M.; Feng, Y.; Li, H.; Wang, Y.; Ma, Y.; Li, M.; An, F.; Guo, H. EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 2015, 163, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ma, B.; He, S.J.; Xiong, Q.; Duan, K.X.; Yin, C.C.; Chen, H.; Lu, X.; Chen, S.Y.; Zhang, J.S. Maoh6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salt tolerance in rice. Plant Physiol. 2015, 169, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.J.; Chen, H.W.; Ma, B.; Zhang, W.K.; Chen, S.Y.; Zhang, J.S. The role of ethylene in plants under salinity stress. Front. Plant Sci. 2015, 6, 1059. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; do Choi, Y. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Anjum, S.A.; Wang, L.; Farooq, M.; Khan, I.; Xue, L. Methyl jasmonate-induced alteration in lipid peroxidation, antioxidative defence system and yield in soybean under drought. J. Agron. Crop Sci. 2011, 197, 296–301. [Google Scholar] [CrossRef]
- Anjum, S.A.; Tanveer, M.; Hussain, S.; Tung, S.A.; Samad, R.A.; Wang, L.C.; Khan, I.; Rehman, N.U.; Shah, N.; Shahzad, B. Exogenously applied methyl jasmonate improves the drought tolerance in wheat imposed at early and late developmental stages. Acta Physiol. Plant. 2016, 38, 25. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Guo, J.; Zhu, A.; Zhang, L.; Zhang, M. Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicol. Environ. Saf. 2014, 104, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza-sativa L) mediated by agrobacterium and sequence-analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Shon, Y.G.; Lee, S.I.; Kim, C.Y.; Koo, J.C.; Lim, C.O.; Choi, Y.J.; Han, C.D.; Chung, C.H.; Choe, Z.R.; et al. Cultivar variability in the Agrobacterium-rice cell interaction and plant regeneration. Physiol. Plant 1999, 107, 338–345. [Google Scholar] [CrossRef]
- Doyle, P.J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Springer: Berlin/Heidelberg, Germany, 1991; Volume 57, pp. 283–293. [Google Scholar]
- Neff, M.M.; Chory, J. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998, 118, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, M.M. The Effects of Solar UV-B Radiation (280–315 nm) on Higher Plants: Implications of Stratospheric Ozone Reduction. In Research in Photobiology; Plenum Press: New York, NY, USA, 1977; pp. 597–607. [Google Scholar]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. AgriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | log2 (RPKMOXR/RPKMWT) | Description | ||
|---|---|---|---|---|
| 0 h | 6 h | 24 h | ||
| Os02t0575500-01 | −0.16 | −1.13 * | −0.23 | similar to ABC (ATP-binding cassette) transporter-like |
| Os09t0448200-02 | 1.43 ** | 2.62 ** | 1.75 ** | similar to high-affinity potassium transporter |
| Os02t0106100-01 | −1.65 * | −2.73 * | −1.61 * | similar to fructosyltransferase |
| Gene ID | Log2 (RPKMOXR/RPKMWT) | Description | ||
|---|---|---|---|---|
| 0 h | 1 h | 6 h | ||
| Os07t0669700-01 | −0.09 | 1.01 ** | 1.09 ** | potassium transporter 4 |
| Os02t0106100-01 | −1.91 * | −1.08 * | −0.84 | similar to fructosyltransferase |
| Os03t0316200-01 | −0.31 | −2.83 * | 0.05 | similar to galactinol synthase |
| Os01t0170000-01 | −0.73 | −2.78 * | 0.18 | raffinose synthase family protein |
| Os09t0376800-01 | 0.38 | 0.77 | 1.33 ** | similar to trehalose-6-phosphate synthase (OsTPS11) |
| Os02t0611200-03 | −1.27 * | −1.42 * | −2.11 * | similar to s-adenosylmethionine decarboxylase 2 |
| Gene ID | Log2 (RPKMOXR/RPKMWT) | Description | ||
|---|---|---|---|---|
| 0 h | 6 h | 24 h | ||
| Catalase (CAT) | ||||
| Os06t0727200-01 | 0.79 | 1.36 ** | 0.79 | catalase isozyme B (EC 1.11.1.6) |
| Ascorbate peroxidase (APX) | ||||
| Os04t0602100-01 | −1.77 * | −2.21 * | −1.34 * | l-ascorbate peroxidase |
| Peroxiredoxin (PrxR) | ||||
| Os06t0196300-01 | −1.64 * | −0.49 | 0.77 | similar to peroxiredoxin Q |
| Glutathione S-transferase (GST) | ||||
| Os04t0435500-01 | −0.72 | −1.03 * | −0.15 | glutathione S-transferase, c-terminal-like domain containing protein |
| Os10t0530500-01 | −1.42 * | −1.78 * | −2.0 * | similar to glutathione-S-transferase |
| Peroxidase superfamily protein | ||||
| Os07t0677200-01 | −0.04 | 2.81 ** | 0.64 | peroxidase |
| Os07t0677400-00 | −1.47 * | −0.28 | −0.02 | peroxidase. |
| Os04t0688100-01 | 0.45 | 2.21 ** | 0.18 | peroxidase (EC 1.11.1.7) |
| Os04t0688100-02 | 0.79 | 1.87 ** | 0.58 | peroxidase (EC 1.11.1.7) |
| Os04t0688500-01 | 0.9 | 2.72 ** | 1.21 ** | peroxidase (EC 1.11.1.7) |
| Os10t0109300-01 | 0.35 | 1.94 ** | 0.17 | similar to peroxidase (EC 1.11.1.7) |
| Os01t0263300-01 | 0.44 | 1.42 ** | −0.6 | similar to peroxidase 72 precursor (EC 1.11.1.7) |
| Os01t0962700-01 | 0.47 | 1.11 ** | −0.16 | similar to peroxidase 12 precursor (EC 1.11.1.7) |
| Os01t0963000-01 | 1.01 ** | 4.55 ** | 1.38 ** | similar to peroxidase BP 1 precursor |
| Os06t0547400-01 | 0.73 | 1.76 ** | 1.03 ** | similar to peroxidase P7 (EC 1.11.1.7) |
| Os06t0695500-01 | 0.01 | 2.19 ** | −0.44 | similar to peroxidase 16 precursor (EC 1.11.1.7) |
| Os04t0688300-01 | 1.02 ** | 3.24 ** | 1.77 ** | Haem peroxidase, plant/fungal/bacterial family protein. |
| Os01t0326300-01 | −0.07 | 1.92** | 0.24 | Haem peroxidase, plant/fungal/bacterial family protein. |
| Os06t0521500-01 | 1.26 ** | 3.09 ** | 0.82 | Haem peroxidase family protein. |
| Os06t0306300-01 | 0.42 | 1.94 ** | 0 | Haem peroxidase domain containing protein. |
| Os02t0684400-01 | 0.24 | 1.33 ** | 0 | similar to OXS3 (OXIDATIVE STRESS 3) |
| Dehydrogenase/reductase | ||||
| Os04t0531900-01 | 0.62 | 1.05 ** | −0.07 | short-chain dehydrogenase/reductase SDR domain-containing protein |
| Os11t0484500-01 | 0.31 | −0.76 | −1.23 * | similar to 6-phosphogluconate dehydrogenase |
| Gene ID | log2 (RPKMOXR/RPKMWT) | Description | ||
|---|---|---|---|---|
| 0 h | 1 h | 6 h | ||
| Catalase (CAT) | ||||
| Os02t0115700-01 | −0.09 | 1.6 ** | 0.67 | catalase isozyme A (EC 1.11.1.6) |
| Os06t0727200-01 | −0.46 | 0.76 | 1.04 ** | catalase isozyme B (EC 1.11.1.6) |
| Os06t0727200-02 | 1.64 ** | 0.57 | −0.31 | catalase isozyme B (EC 1.11.1.6) |
| Ascorbate peroxidase (APX) | ||||
| Os04t0602100-01 | −2.23 * | −2.37 * | −1.6 * | l-ascorbate peroxidase |
| Os06t0567900-01 | 0.67 | 1.15 ** | −0.01 | similar to ascorbate oxidase (Fragment) |
| Peroxiredoxin (PrxR) | ||||
| Os06t0196300-01 | −1.01 * | −0.62 | −0.64 | similar to peroxiredoxin Q (Fragment) |
| Os06t0196300-02 | 0.04 | −1.02 * | −0.07 | similar to peroxiredoxin Q (Fragment) |
| Glutathione S-transferase (GST) | ||||
| Os01t0369700-01 | −0.77 | 0.61 | 1.26 ** | similar to glutathione S-transferase GST 8 |
| Os04t0435500-01 | −0.38 | −1.88 * | −0.26 | glutathione S-transferase, C-terminal-like domain containing protein |
| Peroxidase (POD) | ||||
| Os04t0688100-02 | 0.77 | 0.83 | 2.95 ** | peroxidase (EC 1.11.1.7) |
| Os04t0688100-01 | −0.1 | 1.36 ** | 0.18 | peroxidase (EC 1.11.1.7) |
| Os04t0688500-01 | 0.44 | −0.09 | 1.07 ** | peroxidase (EC 1.11.1.7) |
| Os07t0677400-00 | −1.08 * | −3.03 * | −0.37 | peroxidase |
| Os03t0121200-01 | −0.25 | −2.39 * | −0.67 | similar to Peroxidase 1 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dou, M.; Fan, S.; Yang, S.; Huang, R.; Yu, H.; Feng, X. Overexpression of AmRosea1 Gene Confers Drought and Salt Tolerance in Rice. Int. J. Mol. Sci. 2017, 18, 2. https://doi.org/10.3390/ijms18010002
Dou M, Fan S, Yang S, Huang R, Yu H, Feng X. Overexpression of AmRosea1 Gene Confers Drought and Salt Tolerance in Rice. International Journal of Molecular Sciences. 2017; 18(1):2. https://doi.org/10.3390/ijms18010002
Chicago/Turabian StyleDou, Mingzhu, Sanhong Fan, Suxin Yang, Rongfeng Huang, Huiyun Yu, and Xianzhong Feng. 2017. "Overexpression of AmRosea1 Gene Confers Drought and Salt Tolerance in Rice" International Journal of Molecular Sciences 18, no. 1: 2. https://doi.org/10.3390/ijms18010002






