Tumor LINE-1 Methylation Level in Association with Survival of Patients with Stage II Colon Cancer
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
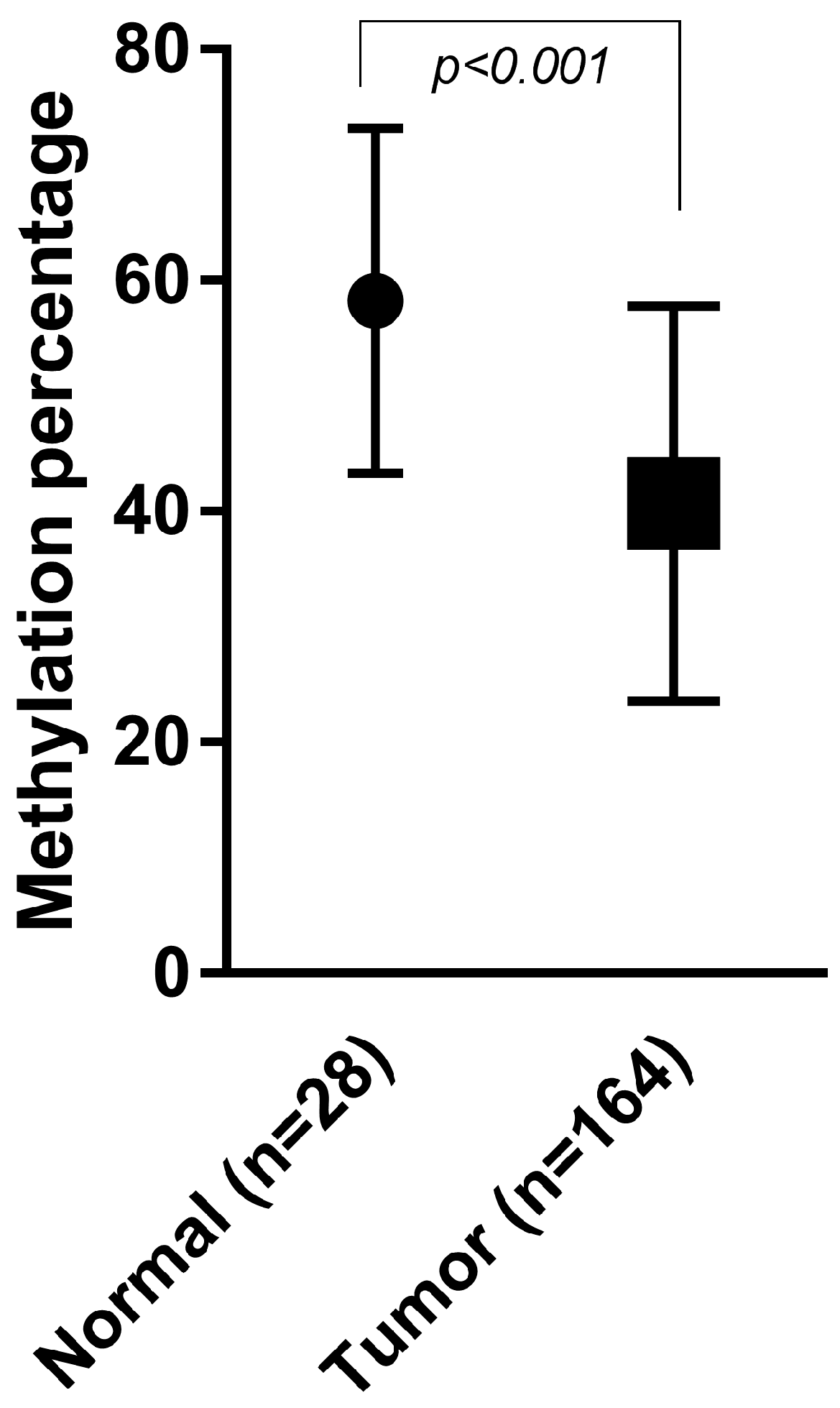
2.2. Methylation Assay Verification
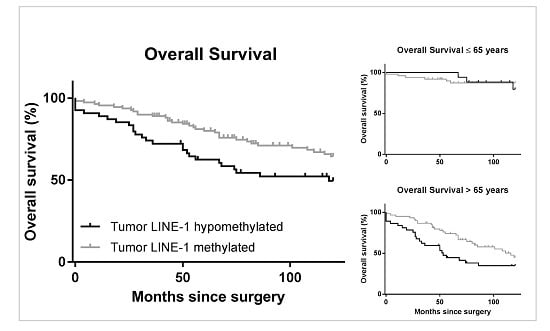
2.3. LINE-1 Methylation Level and Patient Survival
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. DNA Extraction from Formalin-Fixed Paraffin-Embedded Tumor Tissues and Bisulfite Conversion
4.3. Quantitative Real-Time PCR
4.4. Analysis of LINE-1 Methylation Levels
4.5. MLH1 Methylation Status
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| DFS | Disease-free survival |
| HEX | Hexachloro-Fluorescein |
| FAM | 6-Fluorescein amidite |
| FFPE | Formalin-fixed paraffin-embedded |
| LINE-1 | Long interspersed nucleotide element |
| MGB | Minor-groove-binding |
| MSI | Microsatellite instable |
| OS | Overall survival |
| RFS | Recurrence-free survival |
| RFU | Relative fluorescence units |
| (q)PCR | (Quantitative) polymerase chain reaction |
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Goelz, S.E.; Vogelstein, B.; Hamilton, S.R.; Feinberg, A.P. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science 1985, 228, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.S.; Estecio, M.R.; Doshi, K.; Kondo, Y.; Tajara, E.H.; Issa, J.P. A simple method for estimating global DNA methylation using bisulfite pcr of repetitive DNA elements. Nucleic Acids Res. 2004, 32, e38. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Szak, S.T.; Boeke, J.D. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 2004, 429, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Speek, M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 2001, 21, 1973–1985. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Vinciguerra, M.; Agodi, A. LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: A systematic review and meta-analysis. PLoS ONE 2014, 9, e109478. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Nosho, K.; Kirkner, G.J.; Kawasaki, T.; Chan, A.T.; Schernhammer, E.S.; Giovannucci, E.L.; Fuchs, C.S. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J. Natl. Cancer Inst. 2008, 100, 1734–1738. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Huttenhower, C.; Nosho, K.; Tanaka, N.; Shima, K.; Hazra, A.; Schernhammer, E.S.; Hunter, D.J.; Giovannucci, E.L.; Fuchs, C.S.; et al. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol. Cancer 2010, 9, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunami, E.; de Maat, M.; Vu, A.; Turner, R.R.; Hoon, D.S. LINE-1 hypomethylation during primary colon cancer progression. PLoS ONE 2011, 6, e18884. [Google Scholar] [CrossRef] [PubMed]
- Murata, A.; Baba, Y.; Watanabe, M.; Shigaki, H.; Miyake, K.; Ishimoto, T.; Iwatsuki, M.; Iwagami, S.; Sakamoto, Y.; Miyamoto, Y.; et al. Methylation levels of LINE-1 in primary lesion and matched metastatic lesions of colorectal cancer. Br. J. Cancer 2013, 109, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Benard, A.; van de Velde, C.J.; Lessard, L.; Putter, H.; Takeshima, L.; Kuppen, P.J.; Hoon, D.S. Epigenetic status of LINE-1 predicts clinical outcome in early-stage rectal cancer. Br. J. Cancer 2013, 109, 3073–3083. [Google Scholar] [CrossRef] [PubMed]
- Kapiteijn, E.; Liefers, G.J.; Los, L.C.; Kranenbarg, E.K.; Hermans, J.; Tollenaar, R.A.; Moriya, Y.; van de Velde, C.J.; van Krieken, J.H. Mechanisms of oncogenesis in colon versus rectal cancer. J. Pathol. 2001, 195, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Li, J.N.; Zhao, L.; Wu, J.; Wu, B.; Yang, H.; Zhang, H.H.; Qian, J.M. Differences in gene expression profiles and carcinogenesis pathways between colon and rectal cancer. J. Dig. Dis. 2012, 13, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Fransen, K.; Klintenas, M.; Osterstrom, A.; Dimberg, J.; Monstein, H.J.; Soderkvist, P. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis 2004, 25, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Kalady, M.F.; Sanchez, J.A.; Manilich, E.; Hammel, J.; Casey, G.; Church, J.M. Divergent oncogenic changes influence survival differences between colon and rectal adenocarcinomas. Dis. Colon Rectum 2009, 52, 1039–1045. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, N.; Issa, J.P. Aging, methylation and cancer. Histol. Histopathol. 2000, 15, 835–842. [Google Scholar] [PubMed]
- Lao, V.V.; Grady, W.M. Epigenetics and colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Estecio, M.R.; Gharibyan, V.; Shen, L.; Ibrahim, A.E.; Doshi, K.; He, R.; Jelinek, J.; Yang, A.S.; Yan, P.S.; Huang, T.H.; et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS ONE 2007, 2, e399. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Yamauchi, M.; Nishihara, R.; Lochhead, P.; Qian, Z.R.; Kuchiba, A.; Kim, S.A.; Mima, K.; Sukawa, Y.; Jung, S.; et al. Tumor LINE-1 methylation level and microsatellite instability in relation to colorectal cancer prognosis. J. Natl. Cancer Inst. 2014. [Google Scholar] [CrossRef] [PubMed]
- Weisenberger, D.J.; Siegmund, K.D.; Campan, M.; Young, J.; Long, T.I.; Faasse, M.A.; Kang, G.H.; Widschwendter, M.; Weener, D.; Buchanan, D.; et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with braf mutation in colorectal cancer. Nat. Genet. 2006, 38, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Irahara, N.; Nosho, K.; Baba, Y.; Shima, K.; Lindeman, N.I.; Hazra, A.; Schernhammer, E.S.; Hunter, D.J.; Fuchs, C.S.; Ogino, S. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J. Mol. Diagn. 2010, 12, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Nowak, J.A.; Qian, Z.R.; Cao, Y.; Song, M.; Masugi, Y.; Shi, Y.; da Silva, A.; Gu, M.; Li, W.; et al. Tumor LINE-1 methylation level and colorectal cancer location in relation to patient survival. Oncotarget 2016, 7, 55098–55109. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.; Bestor, T.H. Altered methylation patterns in cancer cell genomes: Cause or consequence? Cancer Cell 2002, 1, 299–305. [Google Scholar] [CrossRef]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology 2015, 149, 1204–1225. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Suzuki, I.; Leodolter, A.; Alonso, S.; Horiuchi, S.; Yamashita, K.; Perucho, M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 2006, 9, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Pavicic, W.; Joensuu, E.I.; Nieminen, T.; Peltomaki, P. LINE-1 hypomethylation in familial and sporadic cancer. J. Mol. Med. (Berl.) 2012, 90, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Yamada, A.; Minamiguchi, S.; Sakai, Y.; Horimatsu, T.; Muto, M.; Chiba, T.; Boland, C.R.; Goel, A. Colorectal advanced neoplasms occur through dual carcinogenesis pathways in individuals with coexisting serrated polyps. PLoS ONE 2014, 9, e98059. [Google Scholar] [CrossRef] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Weidner, C.I.; Costa, I.G.; Marioni, R.E.; Ferreira, M.R.; Deary, I.J.; Wagner, W. DNA methylation levels at individual age-associated CPG sites can be indicative for life expectancy. Aging (Albany NY) 2016, 8, 394–401. [Google Scholar] [PubMed]
- Marioni, R.E.; Shah, S.; McRae, A.F.; Chen, B.H.; Colicino, E.; Harris, S.E.; Gibson, J.; Henders, A.K.; Redmond, P.; Cox, S.R.; et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015, 16, 25. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Thibodeau, S.N.; Hamilton, S.R.; Sidransky, D.; Eshleman, J.R.; Burt, R.W.; Meltzer, S.J.; Rodriguez-Bigas, M.A.; Fodde, R.; Ranzani, G.N.; et al. A national cancer institute workshop on microsatellite instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998, 58, 5248–5257. [Google Scholar] [PubMed]
- Ogino, S.; Chan, A.T.; Fuchs, C.S.; Giovannucci, E. Molecular pathological epidemiology of colorectal neoplasia: An emerging transdisciplinary and interdisciplinary field. Gut 2011, 60, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Iskow, R.C.; McCabe, M.T.; Mills, R.E.; Torene, S.; Pittard, W.S.; Neuwald, A.F.; van Meir, E.G.; Vertino, P.M.; Devine, S.E. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 2010, 141, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J.; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K.; et al. Landscape of somatic retrotransposition in human cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Van Roon, E.H.; Boot, A.; Dihal, A.A.; Ernst, R.F.; van Wezel, T.; Morreau, H.; Boer, J.M. BRAF mutation-specific promoter methylation of FOX genes in colorectal cancer. Clin. Epigenet. 2013, 5, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Total n = 164 (%) | LINE-1 | p-Value | ||
|---|---|---|---|---|
| Hypomethylated a n = 54 (%) | Methylated b n = 110 (%) | |||
| Gender | ||||
| Male | 86 (52.4) | 30 (55.6) | 56 (65.1) | 0.58 |
| Female | 78 (47.6) | 24 (44.4) | 54 (49.1) | |
| Age median | 68.0 (±13.6) | 69.5 (±12.0) | 68.0 (±14.3) | 0.16 |
| Age Groups | ||||
| ≤65 | 66 (40.2) | 17 (31.5) | 49 (44.5) | 0.11 |
| >65 | 98 (59.8) | 37 (68.5) | 61 (55.5) | |
| Grade | ||||
| Good | 32 (19.5) | 11 (20.4) | 21 (19.1) | 0.24 |
| Moderate | 79 (48.2) | 30 (55.6) | 49 (48.2) | |
| Poor | 20 (12.2) | 7 (13.0) | 13 (11.8) | |
| Unknown | 33 (9.8) | 6 (11.1) | 27 (24.5) | |
| Location | ||||
| Right | 74 (45.1) | 22 (40.7) | 52 (47.3) | 0.43 |
| Left | 90 (54.9) | 32 (59.3) | 58 (52.7) | |
| MLH1 Promoter | ||||
| Methylated | 30 (18.3) | 6 (11.1) | 24 (21.8) | 0.10 |
| Unmethylated | 134 (81.7) | 48 (88.9) | 86 (78.2) | |
| Adjuvant Therapy | ||||
| No | 156 (95.1) | 50 (92.6) | 106 (96.4) | 0.29 |
| Yes | 8 (49) | 4 (7.4) | 4 (3.6) | |
| Patients n = 164 | Univariate | Multivariate a | |||
|---|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | ||
| Overall Survival | |||||
| LINE-1 Methylated | 110 | 1.00 (reference) | 0.02 | 1.00 (reference) | 0.04 |
| LINE-1 Hypomethylated | 54 | 1.75 (1.09–2.81) | 1.68 (1.03–2.75) | ||
| Disease-free Survival b | |||||
| LINE-1 Methylated | 110 | 1.00 (reference) | 0.07 | 1.00 (reference) | 0.13 |
| LINE-1 Hypomethylated | 54 | 1.52 (0.97–2.40) | 1.43 (0.90–2.29) | ||
| Recurrence-free Survival c | |||||
| LINE-1 Methylated | 110 | 1.00 (reference) | 0.37 | 1.00 (reference) | 0.33 |
| LINE-1 Hypomethylated | 54 | 1.40 (0.66–2.97) | 1.46 (0.68–3.15) | ||
| Overall Survival by MLH1 Status | |||||
| MLH1 Unmethylated | 134 | ||||
| LINE-1 Methylated | 86 | 1.00 (reference) | 0.02 | 1.00 (reference) | 0.09 |
| LINE-1 Hypomethylated | 48 | 1.85 (1.10–3.11) | 1.56 (0.93–2.63) | ||
| MLH1 Methylated | 30 | ||||
| LINE-1 Methylated | 24 | 1.00 (reference) | 0.89 | - | - |
| LINE-1 Hypomethylated | 6 | 1.09 (0.29–4.07) | - | ||
| Patients n = 164 | Univariate | Multivariate a | p-Value for Interaction b | |||
|---|---|---|---|---|---|---|
| HR (95%CI) | p-Value | HR (95% CI) | p-Value | |||
| Overall Survival | ||||||
| ≤65 years | 66 | 0.24 | ||||
| LINE-1 Methylated | 49 | 1.00 (reference) | 0.82 | 1.00 (reference) | 0.80 | |
| LINE-1 Hypomethylated | 17 | 0.81 (0.22–3.04) | 0.84 (0.23–3.16) | |||
| >65 years | 98 | |||||
| LINE-1 Methylated | 61 | 1.00 (reference) | 0.01 | 1.00 (reference) | 0.02 | |
| LINE-1 Hypomethylated | 37 | 1.93 (1.15–3.26) | 2.00 (1.13–3.52) | |||
| Disease-Free Survival c | ||||||
| ≤65 years | 66 | 0.31 | ||||
| LINE-1 Methylated | 49 | 1.00 (reference) | 0.76 | 1.00 (reference) | 0.71 | |
| LINE-1 Hypomethylated | 17 | 0.84 (0.27–2.56) | 0.71 (0.26–2.53) | |||
| >65 years | 98 | |||||
| LINE-1 Methylated | 61 | 1.00 (reference) | 0.05 | 1.00 (reference) | 0.04 | |
| LINE-1 Hypomethylated | 37 | 1.67 (1.00–2.77) | 1.76 (1.02–3.05) | |||
| Recurrence-Free Survival | ||||||
| ≤65 years | 66 | 0.35 | ||||
| LINE-1 Methylated | 49 | 1.00 (reference) | 0.93 | 1.00 (reference) | 0.99 | |
| LINE-1 Hypomethylated | 17 | 0.94 (0.25–3.55) | 1.00 (0.26–3.08) | |||
| >65 years | 98 | |||||
| LINE-1 Methylated | 61 | 1.00 (reference) | 0.28 | 1.00 (reference) | 0.11 | |
| LINE-1 Hypomethylated | 37 | 1.67 (0.66–4.23) | 2.32 (0.83–6.52) | |||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swets, M.; Zaalberg, A.; Boot, A.; Van Wezel, T.; Frouws, M.A.; Bastiaannet, E.; Gelderblom, H.; Van de Velde, C.J.H.; Kuppen, P.J.K. Tumor LINE-1 Methylation Level in Association with Survival of Patients with Stage II Colon Cancer. Int. J. Mol. Sci. 2017, 18, 36. https://doi.org/10.3390/ijms18010036
Swets M, Zaalberg A, Boot A, Van Wezel T, Frouws MA, Bastiaannet E, Gelderblom H, Van de Velde CJH, Kuppen PJK. Tumor LINE-1 Methylation Level in Association with Survival of Patients with Stage II Colon Cancer. International Journal of Molecular Sciences. 2017; 18(1):36. https://doi.org/10.3390/ijms18010036
Chicago/Turabian StyleSwets, Marloes, Anniek Zaalberg, Arnoud Boot, Tom Van Wezel, Martine A. Frouws, Esther Bastiaannet, Hans Gelderblom, Cornelis J. H. Van de Velde, and Peter J. K. Kuppen. 2017. "Tumor LINE-1 Methylation Level in Association with Survival of Patients with Stage II Colon Cancer" International Journal of Molecular Sciences 18, no. 1: 36. https://doi.org/10.3390/ijms18010036