BAG2 Interferes with CHIP-Mediated Ubiquitination of HSP72
Abstract
:1. Introduction
2. Results
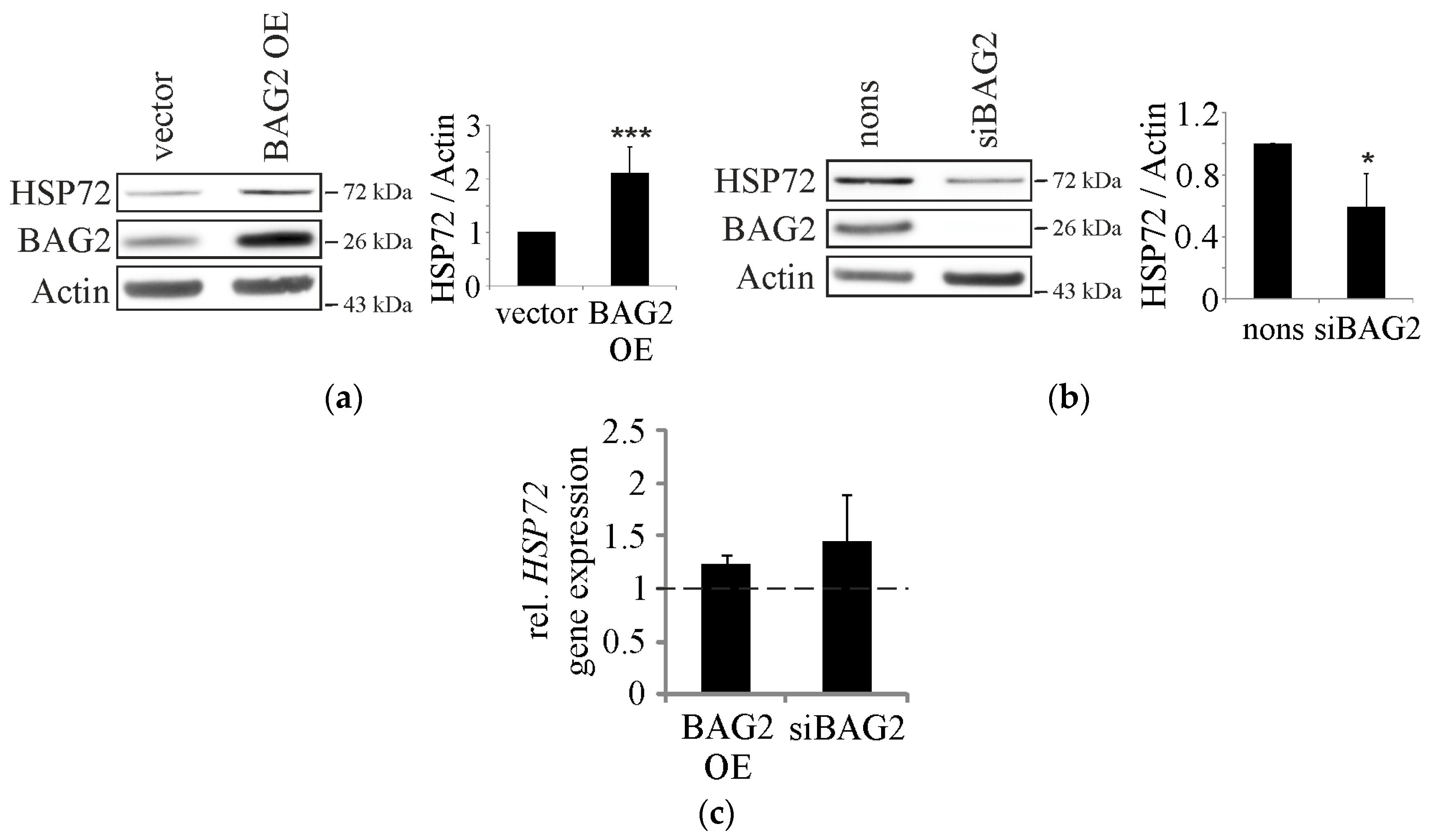
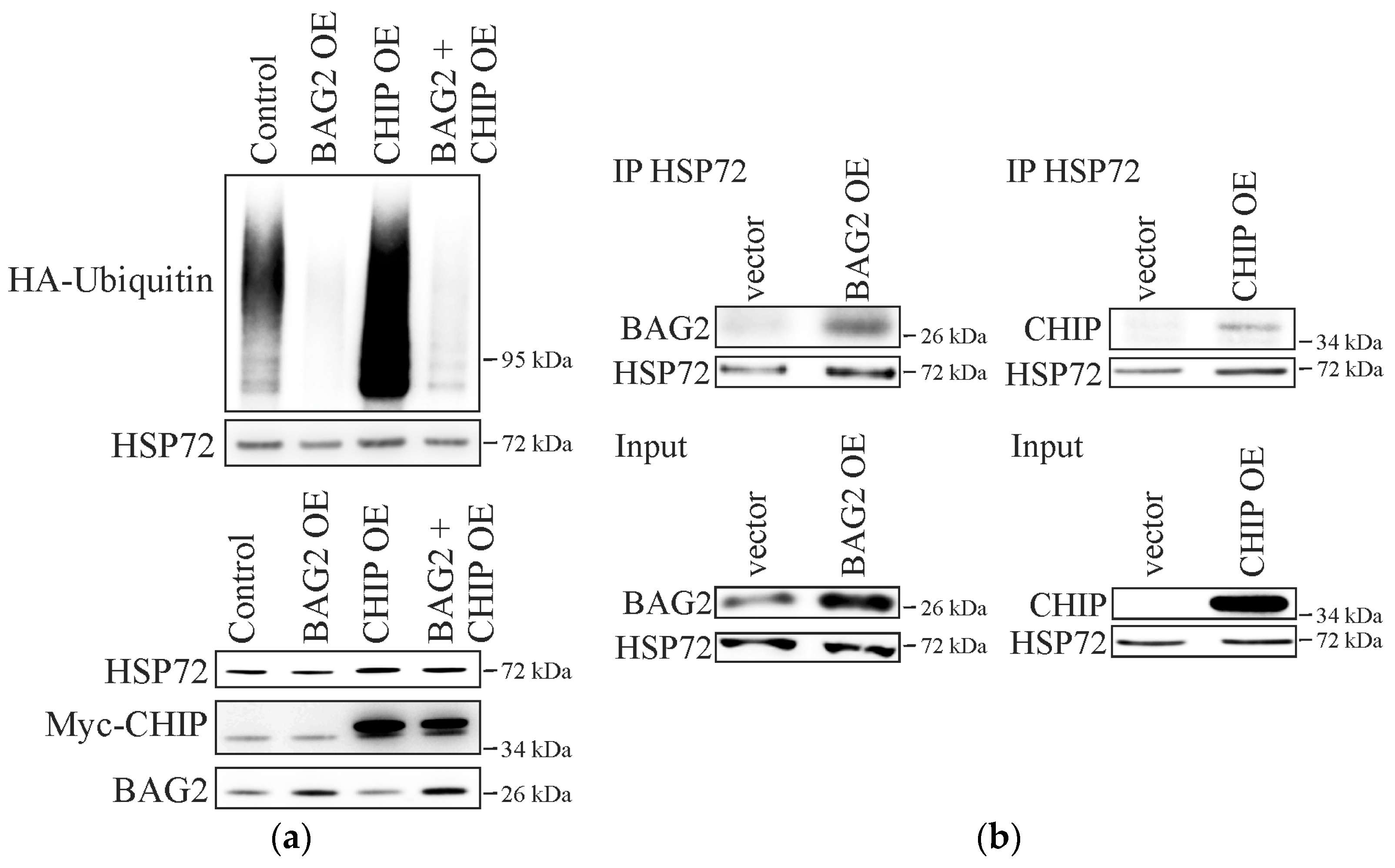
2.1. BAG2 Interferes with CHIP-Mediated Substrate Ubiquitination
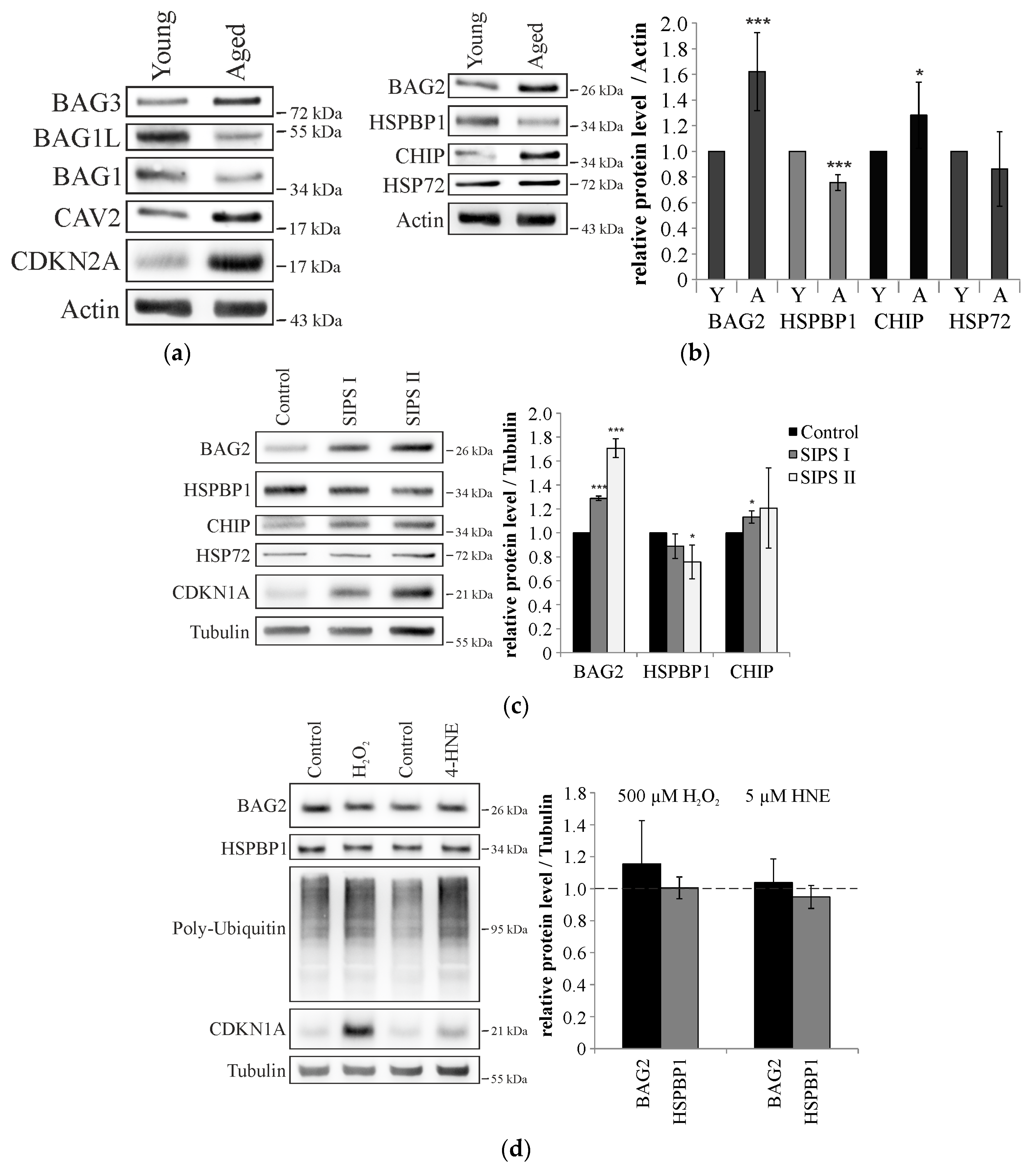
2.2. Cellular Aging Is Associated with Increased CHIP and BAG2 Levels
2.3. BAG2 Inhibits CHIP-Mediated HSP72 Ubiquitination in Aged Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Plasmids, siRNAs and Transfection
4.3. Immunoblotting
4.4. Real-Time PCR
4.5. Co-Immunoprecipitation and Ubiquitination Assay
4.6. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BAG2 | BCL2-associated athanogene 2 |
| CFTR | Cystic fibrosis transmembrane conductance regulator |
| CHIP | C-terminus of HSP70-interacting protein |
| HSP | Heat shock protein |
| HSPBP1 | HSP70 binding protein 1 |
References
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Douglas, P.M.; Dillin, A. Protein homeostasis and aging in neurodegeneration. J. Cell Biol. 2010, 190, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Park, S.H.; Hartl, F.U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014, 24, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Hohfeld, J.; Cyr, D.M.; Patterson, C. From the cradle to the grave: Molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001, 2, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Ballinger, C.A.; Connell, P.; Wu, Y.; Hu, Z.; Thompson, L.J.; Yin, L.Y.; Patterson, C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 1999, 19, 4535–4545. [Google Scholar] [CrossRef] [PubMed]
- Murata, S.; Minami, Y.; Minami, M.; Chiba, T.; Tanaka, K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001, 2, 1133–1138. [Google Scholar] [CrossRef] [PubMed]
- Connell, P.; Ballinger, C.A.; Jiang, J.; Wu, Y.; Thompson, L.J.; Hohfeld, J.; Patterson, C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001, 3, 93–96. [Google Scholar] [PubMed]
- Paul, I.; Ghosh, M.K. A chipotle in physiology and disease. Int. J. Biochem. Cell Biol. 2015, 58, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.; Pandit, L.; Zeng, S.; Eissa, N.T. A critical role for CHIP in the aggresome pathway. Mol. Cell. Biol. 2009, 29, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Min, J.N.; Whaley, R.A.; Sharpless, N.E.; Lockyer, P.; Portbury, A.L.; Patterson, C. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol. Cell. Biol. 2008, 28, 4018–4025. [Google Scholar] [CrossRef] [PubMed]
- Sisoula, C.; Gonos, E.S. CHIP E3 ligase regulates mammalian senescence by modulating the levels of oxidized proteins. Mech. Ageing Dev. 2011, 132, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Bohse, K.; Arndt, V.; Schmitz, A.; Hohfeld, J. The cochaperone HSPBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol. Biol. Cell 2004, 15, 4003–4010. [Google Scholar] [CrossRef] [PubMed]
- Arndt, V.; Daniel, C.; Nastainczyk, W.; Alberti, S.; Hohfeld, J. Bag-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol. Biol. Cell 2005, 16, 5891–5900. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Qian, S.B.; Li, H.H.; McDonough, H.; Borchers, C.; Huang, D.; Takayama, S.; Younger, J.M.; Ren, H.Y.; Cyr, D.M.; et al. Regulation of the cytoplasmic quality control protein degradation pathway by Bag-2. J. Biol. Chem. 2005, 280, 38673–38681. [Google Scholar] [CrossRef] [PubMed]
- Behl, C. Breaking bag: The co-chaperone Bag-3 in health and disease. Trends Pharmacol. Sci. 2016, 37, 672–688. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Sato, T.; Krajewski, S.; Kochel, K.; Irie, S.; Millan, J.A.; Reed, J.C. Cloning and functional analysis of Bag-1: A novel BCL-2-binding protein with anti-cell death activity. Cell 1995, 80, 279–284. [Google Scholar] [CrossRef]
- Doukhanina, E.V.; Chen, S.; van der Zalm, E.; Godzik, A.; Reed, J.; Dickman, M.B. Identification and functional characterization of the bag protein family in arabidopsis thaliana. J. Biol. Chem. 2006, 281, 18793–18801. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Reed, J.C. Molecular chaperone targeting and regulation by bag family proteins. Nat. Cell Biol. 2001, 3, E237–E241. [Google Scholar] [CrossRef] [PubMed]
- Sisoula, C.; Trachana, V.; Patterson, C.; Gonos, E.S. CHIP-dependent p53 regulation occurs specifically during cellular senescence. Free Radic. Biol. Med. 2011, 50, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J.; d’Adda di Fagagna, F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007, 8, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Gamerdinger, M.; Hajieva, P.; Kaya, A.M.; Wolfrum, U.; Hartl, F.U.; Behl, C. Protein quality control during aging involves recruitment of the macroautophagy pathway by Bag-3. EMBO J. 2009, 28, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Kern, A.; Roempp, B.; Prager, K.; Walter, J.; Behl, C. Down-regulation of endogenous amyloid precursor protein processing due to cellular aging. J. Biol. Chem. 2006, 281, 2405–2413. [Google Scholar] [CrossRef] [PubMed]
- Toussaint, O.; Medrano, E.E.; von Zglinicki, T. Cellular and molecular mechanisms of stress-induced premature senescence (SIPS) of human diploid fibroblasts and melanocytes. Exp. Gerontol. 2000, 35, 927–945. [Google Scholar] [CrossRef]
- Qian, S.B.; McDonough, H.; Boellmann, F.; Cyr, D.M.; Patterson, C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and HSP70. Nature 2006, 440, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Morawe, T.; Hiebel, C.; Kern, A.; Behl, C. Protein homeostasis, aging and Alzheimer’s disease. Mol. Neurobiol. 2012, 46, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Luders, J.; Demand, J.; Hohfeld, J. The ubiquitin-related Bag-1 provides a link between the molecular chaperones Hsc70/HSP70 and the proteasome. J. Biol. Chem. 2000, 275, 4613–4617. [Google Scholar] [CrossRef] [PubMed]
- Spang, N.; Feldmann, A.; Huesmann, H.; Bekbulat, F.; Schmitt, V.; Hiebel, C.; Koziollek-Drechsler, I.; Clement, A.M.; Moosmann, B.; Jung, J.; et al. RAB3GAP1 and RAB3GAP2 modulate basal and rapamycin-induced autophagy. Autophagy 2014, 10, 2297–2309. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schönbühler, B.; Schmitt, V.; Huesmann, H.; Kern, A.; Gamerdinger, M.; Behl, C. BAG2 Interferes with CHIP-Mediated Ubiquitination of HSP72. Int. J. Mol. Sci. 2017, 18, 69. https://doi.org/10.3390/ijms18010069
Schönbühler B, Schmitt V, Huesmann H, Kern A, Gamerdinger M, Behl C. BAG2 Interferes with CHIP-Mediated Ubiquitination of HSP72. International Journal of Molecular Sciences. 2017; 18(1):69. https://doi.org/10.3390/ijms18010069
Chicago/Turabian StyleSchönbühler, Bianca, Verena Schmitt, Heike Huesmann, Andreas Kern, Martin Gamerdinger, and Christian Behl. 2017. "BAG2 Interferes with CHIP-Mediated Ubiquitination of HSP72" International Journal of Molecular Sciences 18, no. 1: 69. https://doi.org/10.3390/ijms18010069




