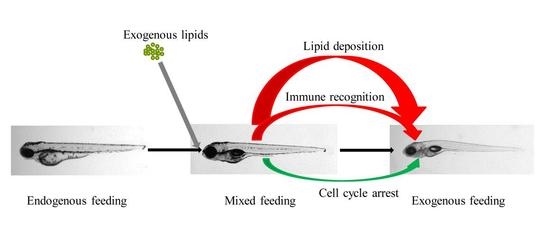
Transcriptome Analysis Reveals Increases in Visceral Lipogenesis and Storage and Activation of the Antigen Processing and Presentation Pathway during the Mouth-Opening Stage in Zebrafish Larvae
Abstract
:1. Introduction
2. Results
2.1. Mapping of RNA-Seq Reads to the Zebrafish Genome
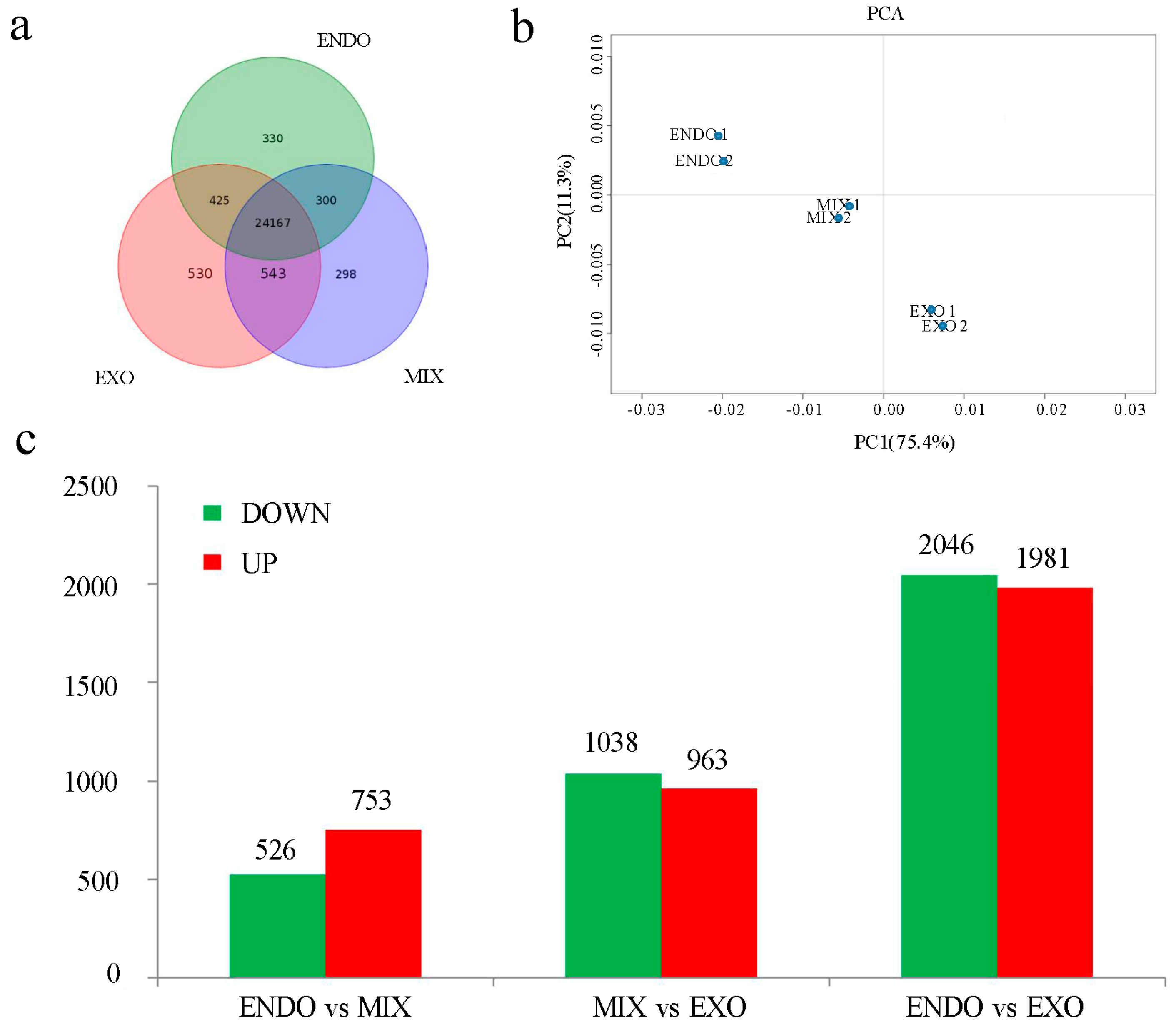
2.2. Gene Expression Detected by RNA-Seq
2.3. Analysis of Differential Expression
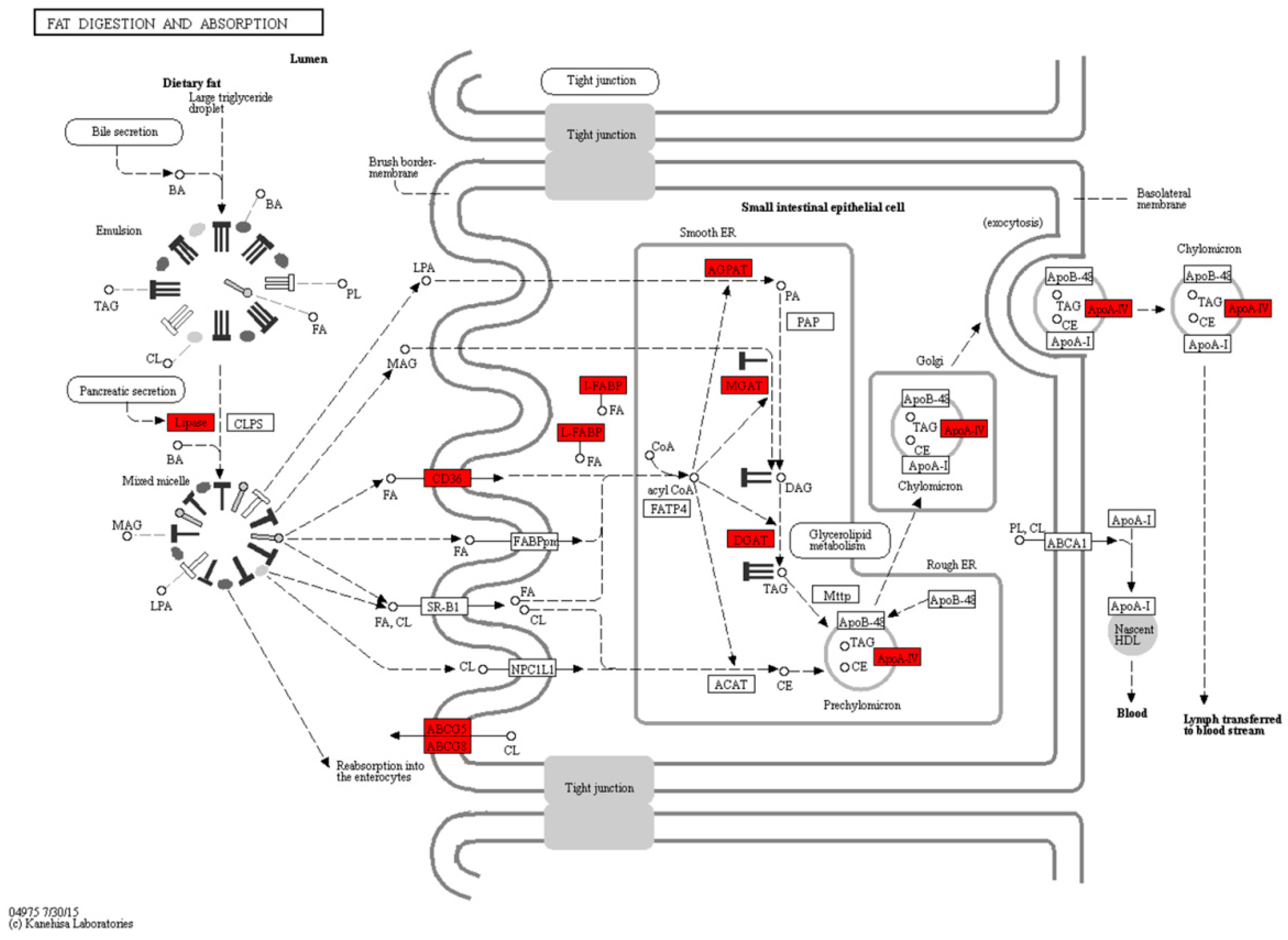
2.4. Fat Digestion, Absorption and Fatty Acid Metabolism during the Mouth-Opening Stage
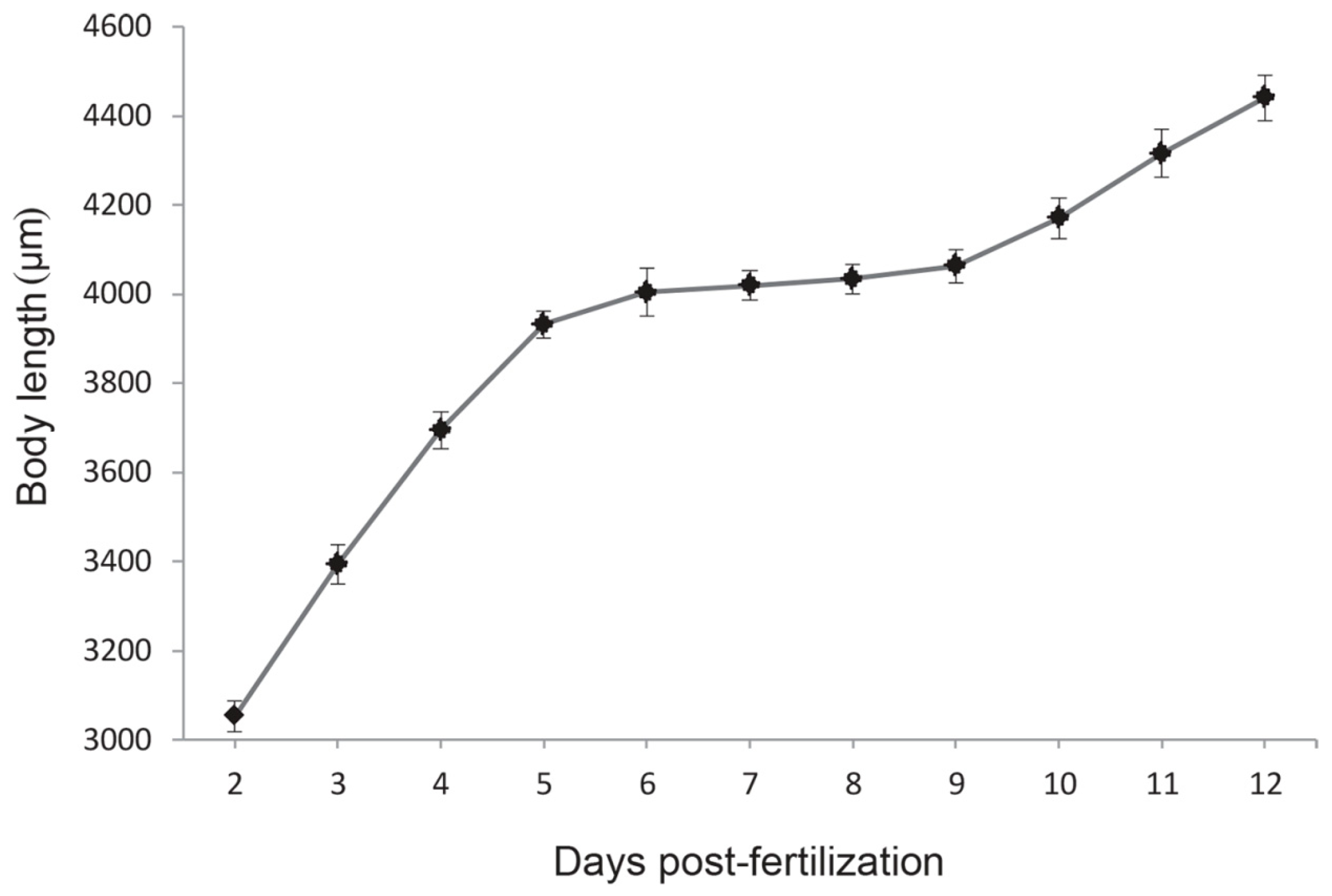
2.5. Temporal Expression of the Growth Hormone and Insulin-Like Growth Factor-I (GH-IGF-I) System and Growth Pattern during the Mouth-Opening Stage
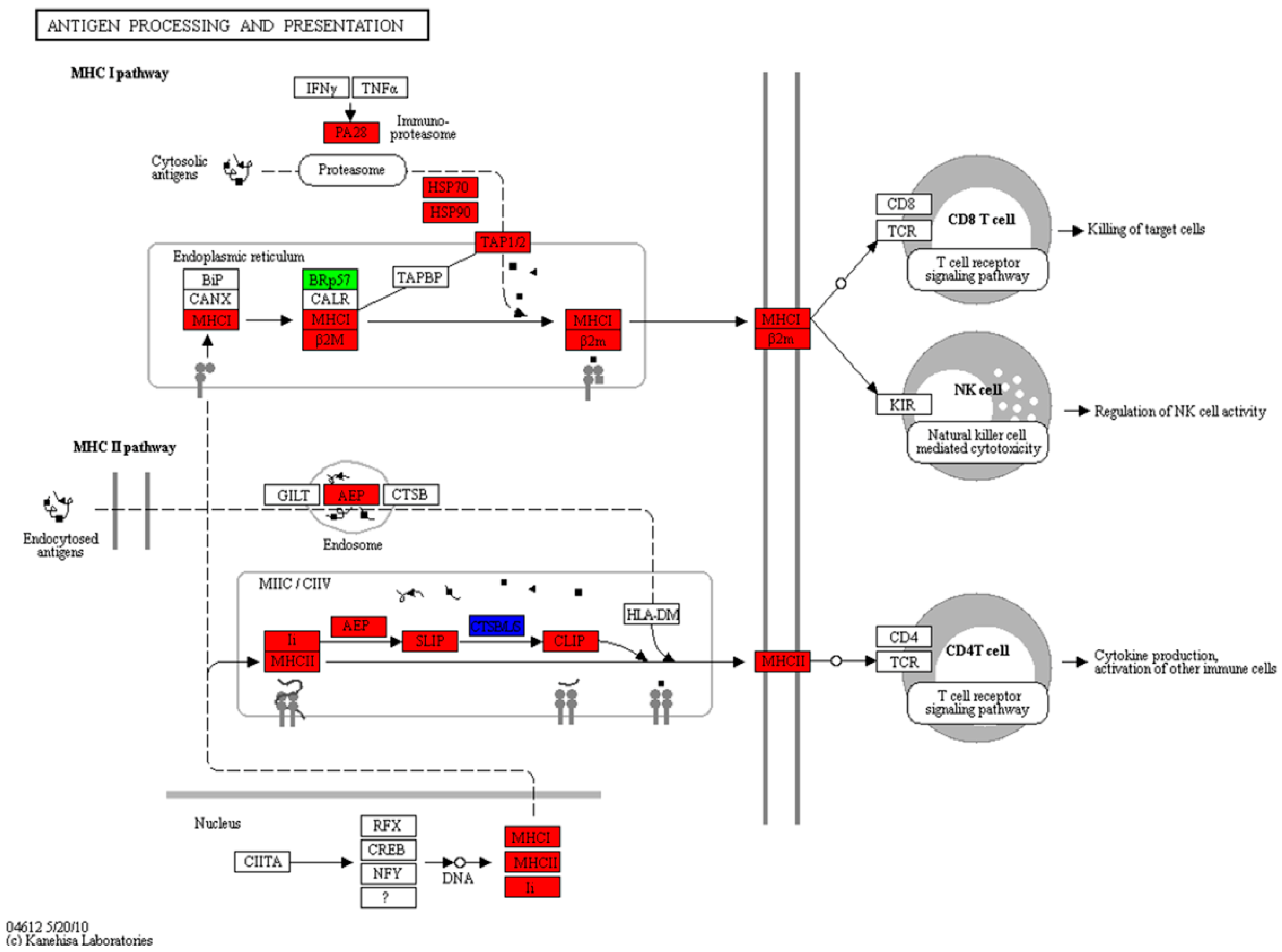
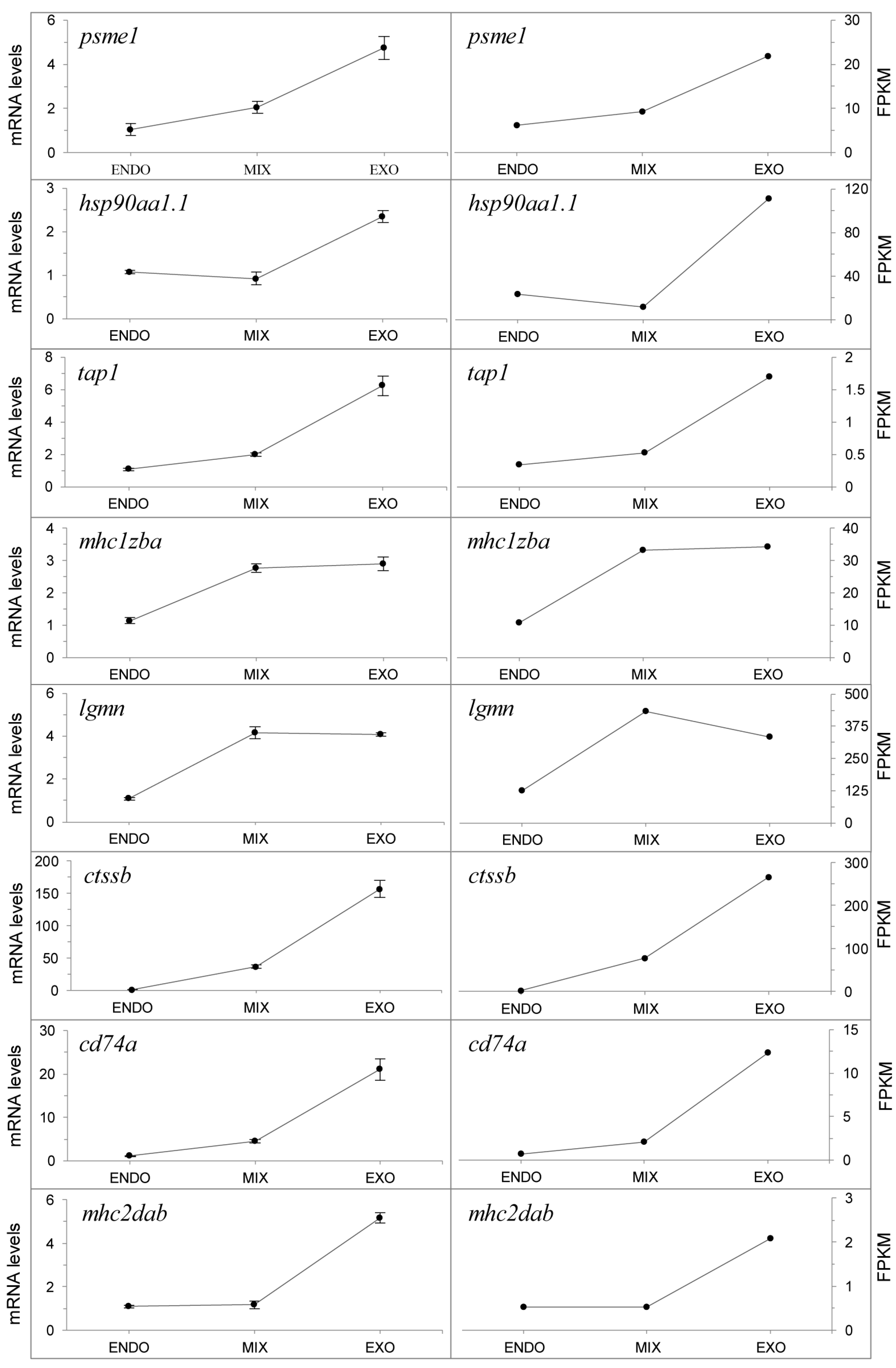
2.6. Expression of the Antigen Processing and Presentation Pathway during the Mouth-Opening Stage
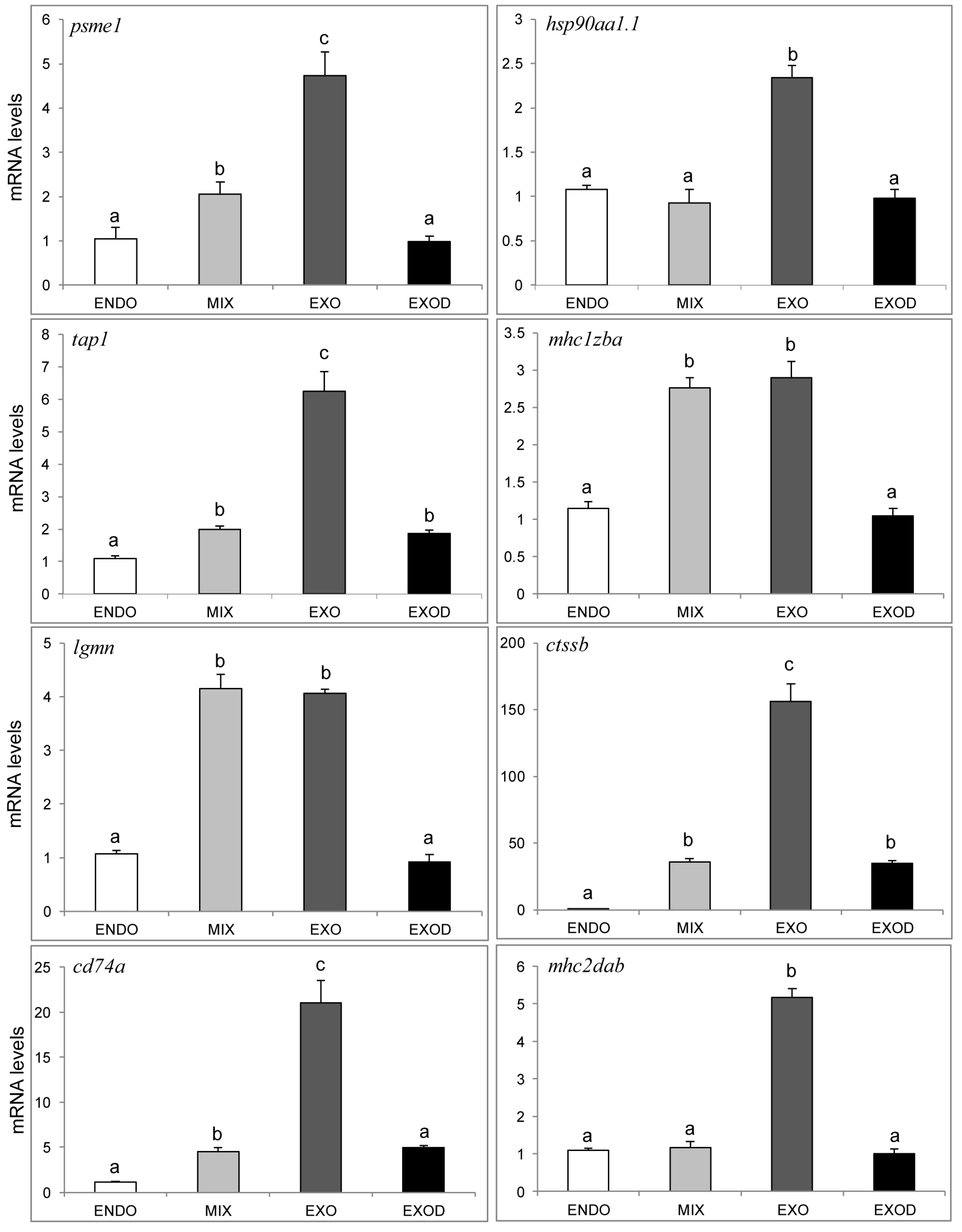
2.7. qPCR Analysis of Genes in the Antigen Processing and Presentation Pathway
3. Discussion
3.1. Exogenous Fatty Acids Are Used for Lipid Deposition Instead of Metabolism during the Mouth-Opening Stage
3.2. Zebrafish Larvae Growth Is Retarded by the Cell Cycle Pathway during the Mouth-Opening Stage
3.3. Activation of the MHC Pathway Depends on the Supply of Exogenous Nutrients during the Mouth-Opening Stage
4. Materials and Methods
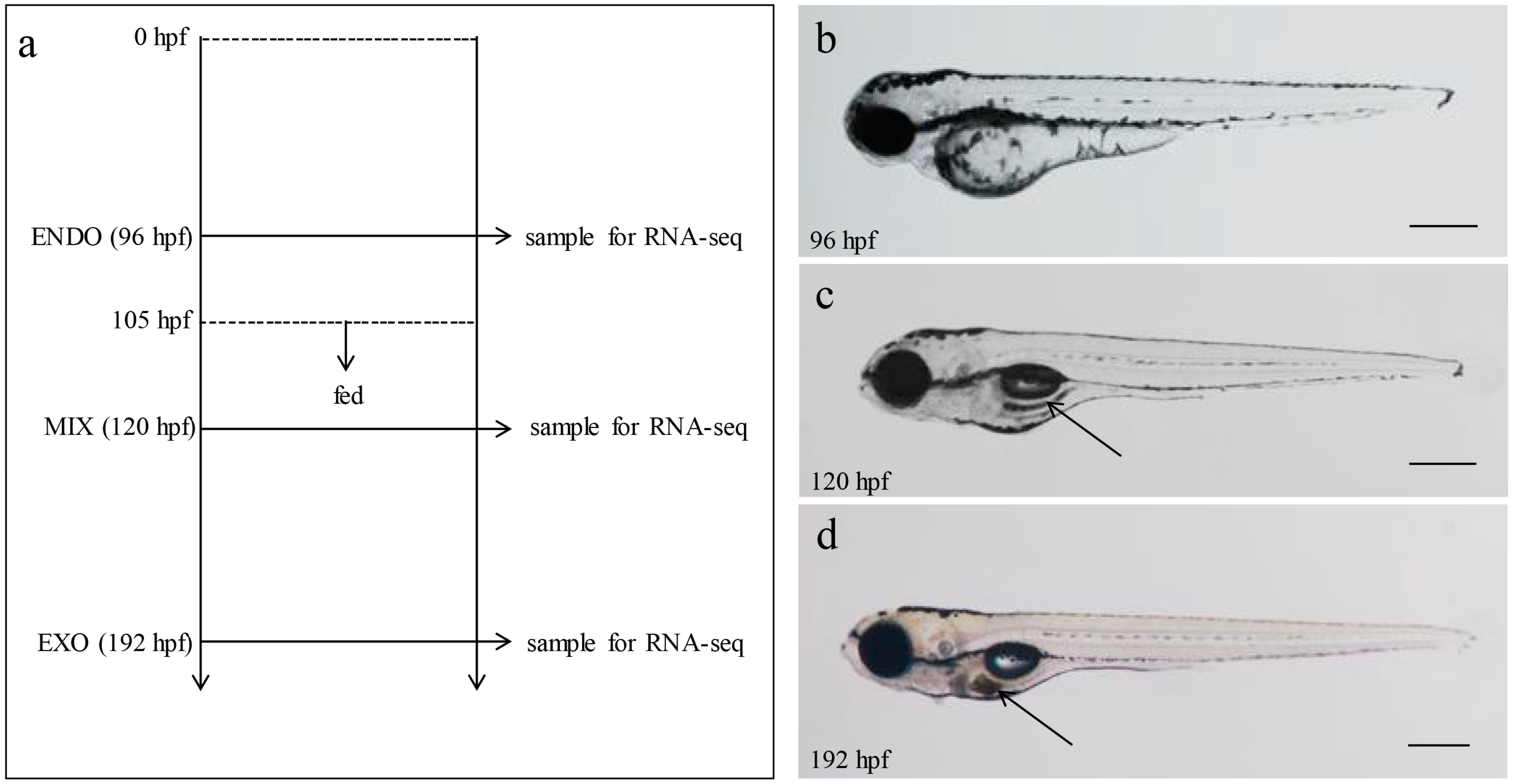
4.1. Larvae Collection and RNA Extraction
4.2. cDNA Library Construction and Illumina Deep Sequencing
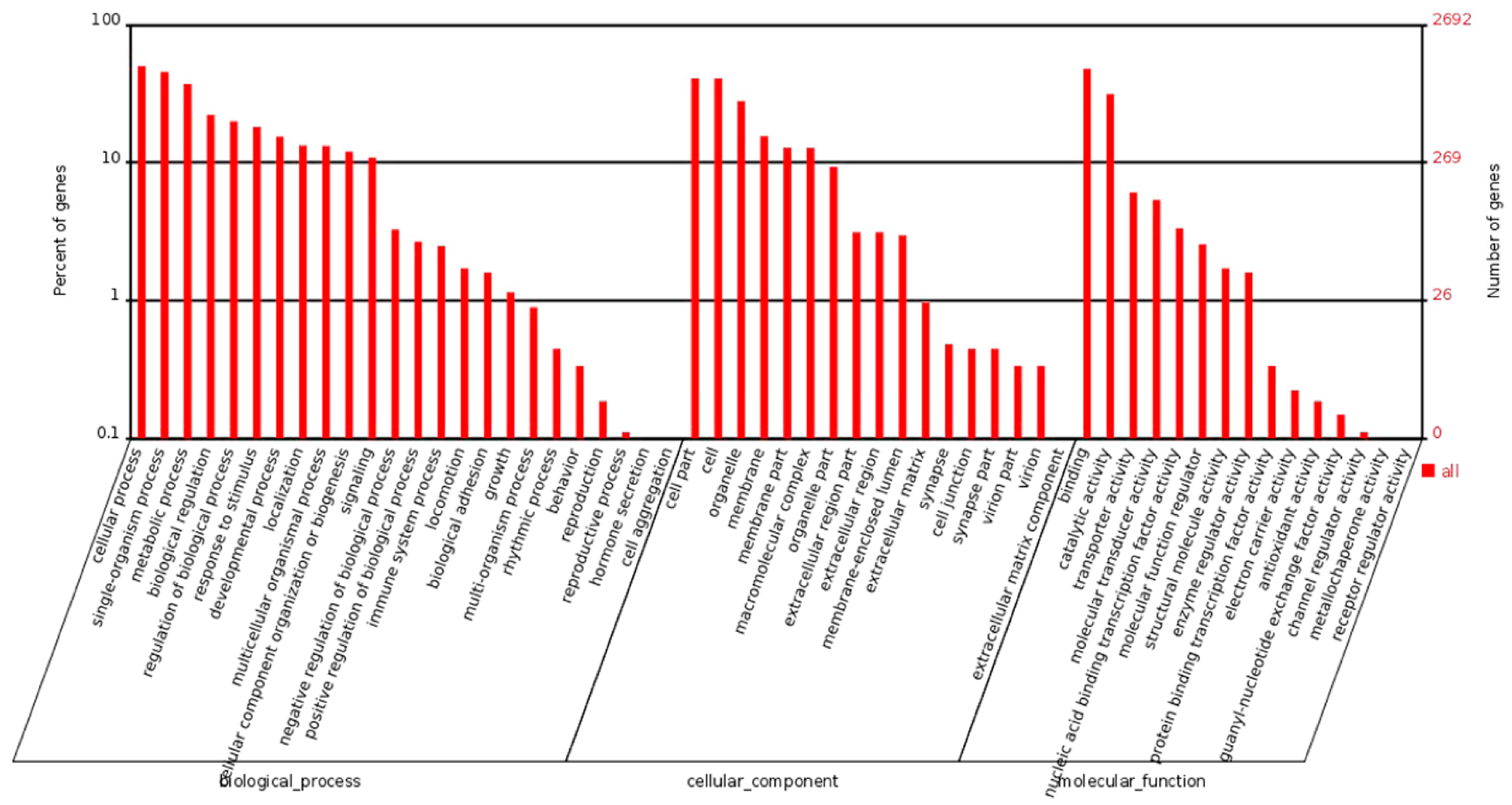
4.3. Bioinformatic Analysis of RNA-Seq Data
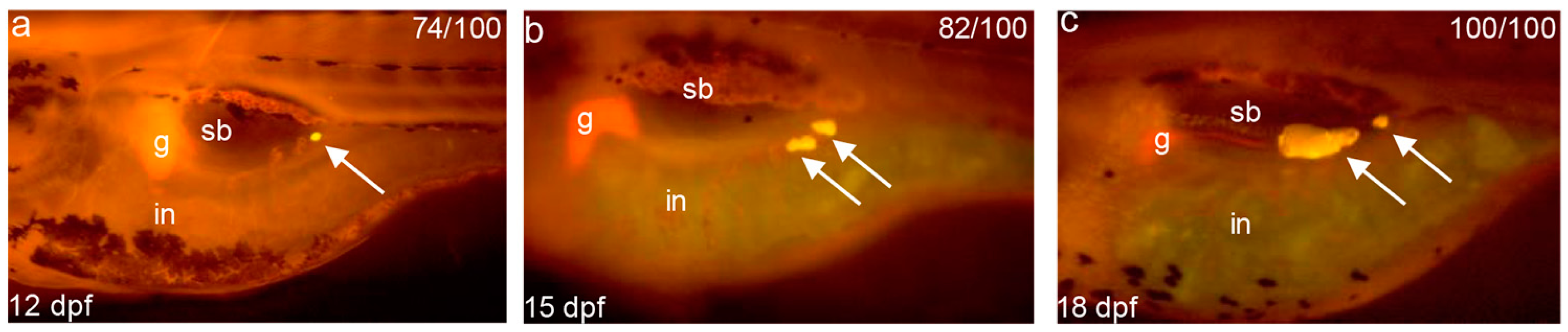
4.4. Zebrafish Larvae Adipocytes Staining and Measure of Body Length
4.5. qPCR Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| α | Alpha |
| β | Beta |
| DEGs | Differentially expressed genes |
| dpf | Days post fertilization |
| ENDO | Endogenous feeding |
| EXO | Exogenous feeding |
| FABP | Fatty acid binding protein |
| FDR | False discovery rate |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| GO | Gene ontology |
| hpf | Hours post fertilization |
| ID | Identification |
| KEGG | Kyoto encyclopedia of genes and genomes |
| MHC | Major histocompatibility complex |
| MIX | Mixed feeding |
| PCA | Principal component analysis |
| PCs | Principal components |
| qPCR | Quantitative real-time polymerase chain reaction |
| RIN | RNA integrity number |
| SRA | Sequence read archive |
| STEM | Short time-series expression miner software |
| TAG | Triglycerides |
| WEGO | Web gene ontology annotation plot |
References
- Mazurais, D.; Darias, M.; Zambonino-infantej, J.L.; Cahuc, C.L. Transcriptomics for understanding marine fish larval development. Can. J. Zool. 2011, 89, 599–611. [Google Scholar] [CrossRef]
- Mennigen, J.A.; Skiba-Cassy, S.; Panserat, S. Ontogenetic expression of metabolic genes and microRNAs in rainbow trout alevins during the transition from the endogenous to the exogenous feeding period. J. Exp. Biol. 2013, 216, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Ahlstrom, E.H.; Amaoka, K.; Hensley, D.A.; Moser, H.G.; Sumida, B.Y. Pleuronectiformes: Development, ontogeny and systematics of fishes. Am. Soc. Ichthyol. Herpetol. Ser. 1984, 1, 640–670. [Google Scholar]
- Andrew, G.J.; Rawlinson, K.A.; Justin, B.; Warrick, S.L.; Clare, V.H.; Baker, C.V.; Shubin, N.H. Holocephalan embryos provide evidence for gill arch appendage reduction and opercular evolution in cartilaginous fishes. Proc. Natl. Acad. Sci. USA 2011, 108, 1507–1512. [Google Scholar]
- Rosenberger, L.J. Pectoral fin locomotion in batoid fishes: Undulation versus oscillation. J. Exp. Biol. 2001, 204, 379–394. [Google Scholar] [PubMed]
- Mascarello, F.; Rowlerson, A.; Veggetti, A. Hyperplasia of lateral muscle during normal growth of the sea-bream, Sparus aurata. J. Muscle Res. Cell Motil. 1994, 15, 189. [Google Scholar]
- Barak, Y.; Nelson, M.C.; Ong, E.S.; Jones, Y.Z.; Ruiz-Lozano, P.; Chien, K.R.; Koder, A.; Evans, R.M. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell 1999, 4, 585–595. [Google Scholar] [CrossRef]
- Sarropoulou, E.; Kotoulas, G.; Power, D.M.; Geisler, R. Gene expression profiling of gilthead sea bream during early development and detection of stress-related genes by application of cDNA microarray technology. Physiol. Genom. 2005, 23, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Rawls, J.F.; Samuel, B.S.; Gordon, J.I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA 2004, 101, 4596–4601. [Google Scholar] [CrossRef] [PubMed]
- Suau, A.; Bonnet, R.; Sutren, M.; Godon, J.J.; Gibson, G.R.; Collins, M.D.; Dore, J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 1999, 65, 4799–4807. [Google Scholar] [PubMed]
- Stappenbeck, T.S.; Hooper, L.V.; Gordon, J.I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 2002, 99, 15451–15455. [Google Scholar] [CrossRef] [PubMed]
- Bry, L.; Falk, P.G.; Midtvedt, T.; Gordon, J.I. A model of host-microbial interactions in an open mammalian ecosystem. Science 1996, 273, 1380–1383. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.; Campbell, J.I.; King, T.P.; Grant, G.; Jansson, E.A.; Coutts, A.G.; Pettersson, S.; Conway, S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat. Immunol. 2004, 5, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Midtvedt, T.; Gordon, J.I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 2002, 22, 283–307. [Google Scholar] [CrossRef] [PubMed]
- Ferraresso, S.; Bonaldo, A.; Parma, L.; Cinotti, S.; Massi, P.; Bargelloni, L.; Gatta, P.P. Exploring the larval transcriptome of the common sole (Soleasolea L.). BMC Genom. 2013, 14, 315. [Google Scholar] [CrossRef] [PubMed]
- Zambonino-Infante, J.L.; Cahu, C.L. Dietary modulation of some digestive enzymes and metabolic processes in developing marine fish: Applications to diet formulation. Aquaculture 2007, 268, 98–105. [Google Scholar] [CrossRef]
- Alestrom, P.; Holter, J.L.; Nourizadeh-Lillabadi, R. Zebrafish in functional genomics and aquatic biomedicine. Trends Biotechnol. 2006, 24, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Detrich, H.W.; Westerfield, M.; Zon, L.I. Overview of the Zebrafish system. Methods Cell Biol. 1999, 59, 3–10. [Google Scholar] [PubMed]
- Li, Y.; Xiang, J.H.; Duan, C. Insulin-like growth factor-binding protein-3 plays an important role in regulating pharyngeal skeleton and inner ear formation and differentiation. J. Biol. Chem. 2005, 280, 3613–3620. [Google Scholar] [CrossRef] [PubMed]
- Paw, B.H.; Zon, L.I. Zebrafish: A genetic approach in studying hematopoiesis. Curr. Opin. Hematol. 2000, 7, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Van den Belt, K.; Verheyen, R.; Witters, H. Comparison of vitellogenin responses in zebrafish and rainbow trout following exposure to environmental estrogens. Ecotoxicol. Environ. Saf. 2003, 56, 271–281. [Google Scholar] [CrossRef]
- Ferraresso, S.; Vitulo, N.; Mininni, A.N.; Romualdi, C.; Cardazzo, B.; Negrisolo, E.; Reinhardt, R.; Canario, A.V.; Patarnello, T.; Bargelloni, L. Development and validation of a gene expression oligo microarray for the gilthead sea bream (Sparus aurata). BMC Genom. 2008, 9, 580. [Google Scholar] [CrossRef] [PubMed]
- Lanes, C.F.; Bizuayehu, T.T.; de Oliveria Fernandes, J.M.; Kiron, V.; Babiak, I. Transcriptome of Altantic cod (Gadus morhua L.) early embryos from farmed and wild broodstocks. Mar. Biotechnol. 2013, 15, 677–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, C.; Zhang, C.F.; Zhang, R.Y.; Zhao, K. Transcriptome profiling analysis of naked carp (Gymnocypris przewalskii) provides insights into the immune-related genes in highland fish. Fish Shellfish Immunol. 2015, 46, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.F.; Wu, Z.H.; Wang, L.J.; Zou, Y.X.; Zhang, P.J.; You, F. Characterization of embryo transcriptome of gynogenetic olive flounder Paralichthys olivaceus. Mar. Biotechnol. 2016, 18, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Zarski, D.; Nguyen, T.; Le Cam, A.; Montfort, J.; Dutto, G.; Vidal, M.O.; Fauvel, C.; Bobe, J. Transcriptomic profiling of egg quality in sea bass (Dicentrarchus labrax) sheds light on genes involved in ubiquitination and translation. Mar. Biotechnol. 2017, 19, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. Lipases as practical biocatalysts. Curr. Opin. Chem. Biol. 2002, 16, 145–150. [Google Scholar] [CrossRef]
- Karanth, S.; Denovan-Wright, E.M.; Thisse, C.; Thisse, B.; Wright, J.M. Tandem duplication of the fabp1b gene and subsequent divergence of the tissue-specific. Genome 2009, 52, 985–992. [Google Scholar] [PubMed]
- Mayer, A.N.; Fishman, M.C. nil per os encodes a conserved RNA recognition motif protein required for morphogenesis and cytodifferentiation of digestive organs in zebrafish. Development 2003, 130, 3917–3928. [Google Scholar] [CrossRef] [PubMed]
- Oehlers, S.H.; Flores, M.V.; Okuda, K.S.; Hall, C.J.; Crosier, K.E.; Crosier, P.S. A chemical enterocolitis model in zebrafish larvae that is dependent on microbiota and responsive to pharmacological agents. Dev. Dyn. 2011, 240, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Denovan-Wright, E.M.; Pierce, M.; Sharma, M.K.; Wright, J.M. cDNA sequence and tissue-specific expression of a basic liver-type fatty acid binding protein in adult zebrafish (Danio rerio). Biochim. Biophys. Acta 2000, 1492, 227–232. [Google Scholar] [CrossRef]
- Bernlohr, D.A.; Coe, N.R.; Licata, V.J. Fatty acid trafficking in the adipocyte. Semin. Cell Dev. Biol. 1999, 10, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Galilea, M.; Tapia, P.; Cautivo, K.; Morselli, E.; Cortes, V.A. AGPAT2 deficiency impairs adipogenic differentiation in primary cultured preadipocytes in a non-autophagy or apoptosis dependent mechanism. Biochem. Biophys. Res. Commun. 2015, 467, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.W.; Gao, Y.; Yen, M.I.; Yen, C.L. Intestine-specific deletion of acyl-CoA: Monoacylglycerol acyltransferase (MGAT) 2 protects mice from diet-induced obesity and glucose intolerance. J. Biol. Chem. 2014, 289, 17338–17349. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.J.; Cases, S.; Jensen, D.R.; Chen, H.C.; Sande, E.; Tow, B.; Sanan, D.A.; Raber, J.; Eckel, R.H.; Farese, R.V. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat. Genet. 2000, 25, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Havel, R.J. Lipoproteins and lipid transport. In Advances in Experimental Medicine and Biology; Springer: Berlin, Germany, 1975; Volume 63, pp. 37–59. [Google Scholar]
- Otis, J.P.; Zeituni, E.M.; Thierer, J.H.; Anderson, J.L.; Brown, A.C.; Boehm, E.D.; Cerchione, D.M.; Ceasrine, A.M.; Avraham-Davidi, I.; Temeplhof, H.; et al. Zebrafish as a model for apolipoprotein biology: Comprehensive expression analysis and a role for ApoA-IV in regulating food intake. Dis. Mod. Mech. 2015, 8, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, V.; van Hul, M.; Lijnen, H.R.; Scroyen, I. CD36 promotes adipocyte differentiation and adipogenesis. Biochim. Biophys. Acta 2012, 1820, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Coburn, C.T.; Hajri, T.; Ibrahimi, A.; Abumrad, N.A. Role of CD36 in membrane transport and utilization of long-chain fatty acids by different tissues. J. Mol. Neurosci. 2001, 16, 117–121. [Google Scholar] [CrossRef]
- Kennedy, D.J.; Kuchibhotla, S.; Westfall, K.M.; Silverstein, R.L.; Morton, R.E.; Febbraio, M. A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signaling. Cardiovasc. Res. 2011, 89, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Mcgarry, J.D.; Brown, N.F. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur. J. Biochem. 1997, 244, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Xue, L.; Huang, H. Liver transcriptome analysis of the Large yellow croaker (Larimichthys crocea) during fasting by using RNA-seq. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.; Douglas, A.; Houlihan, D.F.; Secombes, C.J. Starvation alters the liver transcriptome of the innate response in Atlantic salmon (Salmo salar). BMC Genom. 2010, 11, 418–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, J.G.; Heigenhauser, G.J.; Wood, C.M. Lipid oxidation fuels recovery from exhaustive exercise in white muscle of rainbow trout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 282, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Egginton, S.; Cordiner, S.; Skibeck, C. Thermal compensation of peripheral oxygen transport in skeletal muscle of seasonally acclimatized trout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000, 279, 375–388. [Google Scholar]
- Butler, A.A.; Le Roith, D. Control of growth by the somatotrophic axis: Growth hormone and the insulin-like growth factors have related and independent roles. Ann. Rev. Physiol. 2001, 63, 141–164. [Google Scholar] [CrossRef] [PubMed]
- Hildahl, J.; Power, D.M.; Bjornsson, B.T.; Einarsdottir, I.E. Involvement of growth hormone-insulin-like growth factor I system in cranial remodeling during halibut metamorphosis as indicated by tissue- and stage-specific receptor gene expression and the presence of growth hormone receptor protein. Cell Tissue Res. 2008, 322, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Kamei, H.; Lu, L.; Jiao, S.; Li, Y.; Gyrup, C.; Laursen, L.S.; Oxvig, C.; Zhou, J.; Duan, C. Duplication and diversification of the hypoxia-inducible IGFBP-1 gene in zebrafish. PLoS ONE 2008, 3, e3091. [Google Scholar] [CrossRef] [PubMed]
- Recondo, A. DNA replication in eukaryotic cells. Biochemistry 2002, 71, 333–374. [Google Scholar]
- Gullov, K.; Friis, J.; Bonven, B. Rate of protein synthesis through the cell cycle of Saccharomyces cerevisiae. Exp. Cell Res. 1981, 136, 295–304. [Google Scholar] [CrossRef]
- Trede, N.S.; Langenau, D.M.; Traver, D.; Look, A.T.; Zon, L.I. The use of zebrafish to understand immunity. Immunity 2004, 20, 367–379. [Google Scholar] [CrossRef]
- Forn-Cuni, G.; Varela, M.; Fernandez-Rodriguez, C.M.; Figueras, A.; Novoa, B. Liver immune responses to inflammatory stimuli in a diet-induced obesity model of zebrafish. J. Endocrinol. 2015, 224, 159–170. [Google Scholar] [CrossRef] [PubMed]
- He, J.G.; Deng, M.; Weng, S.P.; Li, Z.; Zhou, S.Y.; Long, Q.X.; Wang, X.Z.; Chan, S.M. Complete genome analysis of the mandarin fish infectious spleen and kidney necrosis iridovirus. Virology 2001, 291, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, L.; Weng, S.; Huang, Z.; Lu, J.; Lan, D.; Zhong, X.; Yu, X.; Xu, A.; He, J. A zebrafish (Danio rerio) model of infectious spleen and kidney necrosis virus (ISKNV) infection. Virology 2008, 376, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rock, K.L.; Farfan-Arribas, D.J.; Shen, J. Proteases in MHC class I presentation and cross-presentation. J. Immunol. 2010, 184, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Hoorn, T.; Paul, P.; Jongsma, M.L.; Neefjes, J. Routes to manipulate MHC class II antigen presentation. Curr. Opin. Immunol. 2011, 23, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Warren, A.; le Couteur, D.G.; Fraser, R.; Bowen, D.G.; McCaughan, G.W.; Bertolino, P. T lymphocytes interact with hepatocytes through fenestrations in murine liver sinusoidal endothelial cells. Hepatology 2006, 44, 1182–1190. [Google Scholar] [CrossRef] [PubMed]
- Crotzer, V.L.; Blum, J.S. Autophagy and its role in MHC-mediated antigen presentation. J. Immunol. 2009, 182, 3335–3341. [Google Scholar] [CrossRef] [PubMed]
- Kubena, K.S.; McMurray, D.N. Nutrition and the immune system: A review of nutrient-nutrient interactions. J. Am. Diet. Assoc. 1996, 96, 1156–1164. [Google Scholar] [CrossRef]
- Wensveen, F.M.; Vaalentic, S.; Marko, S.; Wensveen, T.T.; Polic, B. Interactions between adipose tissue and the immune system in health and malnutrition. Semin. Immunol. 2015, 27, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio); University of Oregon Press: Eugene, OR, USA, 1995. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genom. Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Fang, B.; Chen, J.; Zhang, X.; Luo, Z.; Huang, L.; Chen, X.; Li, Y. De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweet potato (Ipomoea batatas). BMC Genom. 2010, 11, 726. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Song, G.; Yan, J.; He, X.; Li, Q.; Cui, Z. Transcriptomic characterization of cold acclimation in larval zebrafish. BMC Genom. 2013, 14, 612. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics 2006, 7, 191. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Greenspan, P.; Fowler, S.D. Spectrofluorometric studies of the lipid probe, nile red. J. Lipid Res. 1985, 26, 781–789. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Libraries | ENDO-1 | ENDO-2 | MIX-1 | MIX-2 | EXO-1 | EXO-2 |
|---|---|---|---|---|---|---|
| Raw reads (M) | 21.19 | 24.05 | 22.37 | 24.03 | 26.06 | 25.28 |
| Clean reads (M) | 20.74 | 23.54 | 21.84 | 23.47 | 25.50 | 24.71 |
| Percent clean reads | 97.88 | 97.89 | 97.62 | 97.69 | 97.85 | 97.73 |
| Processed reads | 20.37 | 23.05 | 21.16 | 22.74 | 24.91 | 23.97 |
| Mapped reads (M) | 17.33 | 19.14 | 16.97 | 18.43 | 20.80 | 19.45 |
| Percent mapped | 85.07 | 83.06 | 80.20 | 81.05 | 83.50 | 81.12 |
| Unique mapping (M) | 16.96 | 18.81 | 16.67 | 18.09 | 20.41 | 19.12 |
| Percent uniquely mapped reads a | 97.86 | 98.28 | 98.23 | 98.16 | 98.13 | 98.30 |
| Pathways | No. of DEGs with Pathway Annotation | Pathway Identification (ID) | ||||||
|---|---|---|---|---|---|---|---|---|
| All Profiles | Profile 0 | Profile 1 | Profile 3 | Profile 4 | Profile 6 | Profile 7 | ||
| Antigen processing and presentation | 23 | 2 | 0 | 1 | 4 | 5 | 8 | ko04612 |
| Arachidonic acid metabolism | 19 | 1 | 2 | 1 | 2 | 10 | 3 | ko00590 |
| Metabolic pathways | 246 | 59 | 26 | 37 | 17 | 38 | 51 | ko01100 |
| Cell cycle | 32 | 9 | 2 | 19 | 0 | 1 | 1 | ko04110 |
| Linoleic acid metabolism | 10 | 0 | 0 | 1 | 3 | 4 | 2 | ko00591 |
| Homologous recombination | 13 | 4 | 0 | 8 | 0 | 0 | 0 | ko03440 |
| Fat digestion and absorption | 16 | 0 | 0 | 0 | 2 | 6 | 8 | ko04975 |
| Arginine and proline metabolism | 24 | 4 | 1 | 3 | 3 | 5 | 5 | ko00330 |
| Glutathione metabolism | 18 | 3 | 1 | 6 | 0 | 3 | 3 | ko00480 |
| α-Linolenic acid metabolism | 8 | 0 | 0 | 0 | 1 | 2 | 5 | ko00592 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Liu, E.; Li, Y.; Li, X.; Ding, C. Transcriptome Analysis Reveals Increases in Visceral Lipogenesis and Storage and Activation of the Antigen Processing and Presentation Pathway during the Mouth-Opening Stage in Zebrafish Larvae. Int. J. Mol. Sci. 2017, 18, 1634. https://doi.org/10.3390/ijms18081634
Xu H, Liu E, Li Y, Li X, Ding C. Transcriptome Analysis Reveals Increases in Visceral Lipogenesis and Storage and Activation of the Antigen Processing and Presentation Pathway during the Mouth-Opening Stage in Zebrafish Larvae. International Journal of Molecular Sciences. 2017; 18(8):1634. https://doi.org/10.3390/ijms18081634
Chicago/Turabian StyleXu, Hao, Enxiu Liu, Yun Li, Xiaojie Li, and Chenyu Ding. 2017. "Transcriptome Analysis Reveals Increases in Visceral Lipogenesis and Storage and Activation of the Antigen Processing and Presentation Pathway during the Mouth-Opening Stage in Zebrafish Larvae" International Journal of Molecular Sciences 18, no. 8: 1634. https://doi.org/10.3390/ijms18081634