FLP/FRT Recombination from Yeast: Application of a Two Gene Cassette Scheme as an Inducible System in Plants
Abstract
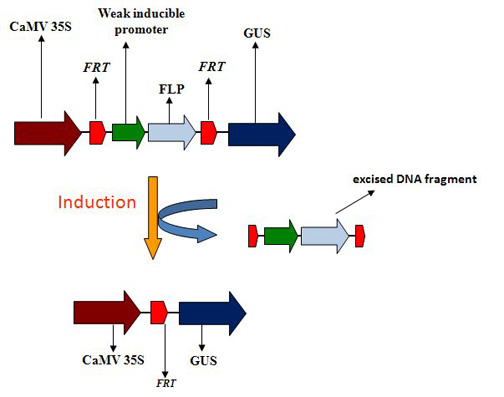
:1. Introduction
2. Experimental Section
2.1. Vector construction
2.2. Plant transformation
2.3. Heat-induction experiments
2.4. GUS assay
2.5. PCR assay
3. Results and Discussion
4. Conclusions
Acknowledgments
References
- Byrne, B; Stack, E; Gilmartin, N; O’ Kennedy, R. Antibody-based sensors: principles, problems and potential for detection of pathogens and associated toxins. Sensors 2009, 9, 4407–4445. [Google Scholar]
- Ferry, N; Gatehouse, AMR. Transgenic crop plants for resistance to biotic stress. In Transgenic Crop Plants; Kole, C, Michler, CH, Abbott, AG, Hall, TC, Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1–65. [Google Scholar]
- Gurr, SJ; Rushton, PJ. Engineering plants with increased disease resistance: How are we going to express it? Trend. Biotech 2005, 23, 283–290. [Google Scholar]
- Stewart, CN, Jr. Monitoring the presence and expression of transgenes in living plants. Trend. Plant Sci 2005, 10, 390–396. [Google Scholar]
- Stewart, CN, Jr. Plant Biotechnology and Genetic: Principles, Techniques, and Applications; Wiley and Sons: Hoboken, NJ, USA, 2008; pp. 1–416. [Google Scholar]
- Kodama, S; Okada, K; Inui, H; Ohkawa, H. Aryl hydrocarbon receptor (AhR)-mediated reporter gene expression systems in transgenic tobacco plants. Planta 2007, 227, 37–45. [Google Scholar]
- Kovalchuk, I; Kovalchuk, O. Transgenic plants as sensors of environmental pollution genotoxicity. Sensors 2008, 8, 1539–1558. [Google Scholar]
- Liew, OW; Chong, PC; Li, B; Asundi, AK. Signature optical cues: emerging technologies for monitoring plant health. Sensors 2008, 8, 3205–3239. [Google Scholar]
- Venter, M; Botha, FC. Synthetic promoter engineering. In Plant Developmental Biology—Biotechnological Perspectives; Pua, EC, Davey, MR, Eds.; Springer: Berlin, Heidelberg, Germany, 2010; Volume 2, pp. 393–414. [Google Scholar]
- Kooshki, M; Mentewab, A; Stewart, CN. Pathogen inducible reporting in transgenic tobacco using a GFP construct. Plant Sci 2003, 165, 213–219. [Google Scholar]
- Mazarei, M; Teplova, I; Hajimorad, MR; Stewart, CN. Pathogen phytosensing: Plants to report plant pathogens. Sensors 2008, 8, 2628–2641. [Google Scholar]
- Ow, DW; Medberry, SL. Genome manipulation through site-specific recombination. CRC Crit. Rev. Plant Sci 1995, 14, 239–261. [Google Scholar]
- Craig, NL. The mechanism of conservative site-specific recombination. Annu. Rev. Genet 1988, 22, 77–105. [Google Scholar]
- Sadowski, PD. Site-specific genetic-recombination—Hops, flips, and flops. FASEB J 1993, 7, 760–767. [Google Scholar]
- Corneille, S; Lutz, K; Svab, Z; Maliga, P. Efficient elimination of selectable marker genes from the plastid genome by the CRE-lox site-specific recombination system. Plant J 2001, 27, 171–178. [Google Scholar]
- Fladung, M; Schenk, TMH; Polak, O; Becker, D. Elimination of marker genes and targeted integration via FLP/FRT recombination system from yeast in hybrid aspen (Populus tremula L. x P. tremuloides Michx.). Tree Genet. Genomes 2009, 6, 205–217. [Google Scholar]
- Gleave, AP; Mitra, DS; Mudge, SR; Morris, BAM. Selectable marker-free transgenic plants without sexual crossing: transient expression of cre recombinase and use of a conditional lethal dominant gene. Plant Mol. Biol 1999, 40, 223–235. [Google Scholar]
- Kausch, AP; Hague, J; Oliver, M; Li, Y; Daniell, H; Maschia, P; Watrud, LS; Stewart, CN, Jr. Transgenic biofuel feedstocks and strategies for bioconfinement. Biofuels 2010, 1, 163–176. [Google Scholar]
- Kilby, NJ; Davies, GJ; Snaith, MR. FLP recombinase in transgenic plants: Constitutive activity in stably transformed tobacco and generation of marked cell clones in Arabidopsis. Plant J 1995, 8, 637–652. [Google Scholar]
- Kilby, NJ; Fyvie, MJ; Sessions, RA; Davies, GJ; Murray, JA. Controlled induction of GUS marked clonal sectors in Arabidopsis. J. Exp. Bot 2000, 51, 853–863. [Google Scholar]
- Lyznik, LA; Gordon-Kamm, WJ; Tao, Y. Site-specific recombination for genetic engineering in plants. Plant Cell Rep 2003, 21, 925–932. [Google Scholar]
- Lyznik, LA; Hirayama, L; Rao, KV; Abad, A; Hodges, TK. Heat-inducible expression of FLP gene in maize cells. Plant J 1995, 8, 177–186. [Google Scholar]
- Luo, H; Lyznik, LA; Gidoni, D; Hodges, TK. FLP-mediated recombination for use in hybrid plant production. Plant J 2000, 23, 423–430. [Google Scholar]
- Luo, K; Sun, M; Deng, W; Xu, S. Excision of selectable marker gene from transgenic tobacco using the GM-gene-deletor system regulated by a heat-inducible promoter. Biotechnol Lett 2008, 30, 1295–1302. [Google Scholar]
- Ow, DW. Recombinase-directed plant transformation for the post-genomic era. Plant Mol. Biol 2002, 48, 183–200. [Google Scholar]
- Srivastava, V; Anderson, OD; Ow, DW. Single-copy transgenic wheat generated through the resolution of complex integration patterns. Proc. Natl. Acad. Sci. USA 1999, 96, 11117–11121. [Google Scholar]
- Srivastava, V; Ow, DW. Single-copy primary transformants of maize obtained through the co-introduction of a recombinase-expressing construct. Plant Mol. Biol 2001, 46, 561–566. [Google Scholar]
- Srivastava, V; Ow, DW. Marker-free site-specific gene integration in plants. Trend. Biotech 2004, 22, 627–629. [Google Scholar]
- Sugita, K; Kasahara, T; Matsunaga, E; Ebinuma, H. A transformation vector for the production of marker-free transgenic plants containing a single copy transgene at high frequency. Plant J 2000, 22, 461–469. [Google Scholar]
- Zuo, JR; Niu, QW; Moller, SG; Chua, NH. Chemical-regulated, site-specific DNA excision in transgenic plants. Nat. Biotech 2001, 19, 157–161. [Google Scholar]
- Clough, SJ; Bent, AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998, 16, 735–743. [Google Scholar]
- Horsch, RB; Fry, JE; Hoffmann, NL; Eichholtz, D; Rogers, SG; Fraley, RT. A simple and general-method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar]
- Jefferson, RA; Kavanagh, TA; Bevan, MW. Gus fusions—beta-glucuronidase as a sensitive and versatile gene fusion marker in higher-plants. EMBO J 1987, 6, 3901–3907. [Google Scholar]
- Cuellar, W; Gaudin, A; Solorzano, D; Casas, A; Nopo, L; Chudalayandi, P; Medrano, G; Kreuze, J; Ghislain, M. Self-excision of the antibiotic resistance gene nptII using a heat inducible Cre-loxP system from transgenic potato. Plant Mol. Biol 2006, 62, 71–82. [Google Scholar]
- Kim, BM; Suehiro, N; Natsuaki, T; Inukai, T; Masuta, C. The P3 protein of turnip mosaic virus can alone induce hypersensitive response-like cell death in Arabidopsis thaliana carrying TuNI. Mol. Plan-Microbe Interact 2010, 23, 144–152. [Google Scholar]




© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Rao, M.R.; Moon, H.S.; Schenk, T.M.H.; Becker, D.; Mazarei, M.; Stewart, C.N., Jr. FLP/FRT Recombination from Yeast: Application of a Two Gene Cassette Scheme as an Inducible System in Plants. Sensors 2010, 10, 8526-8535. https://doi.org/10.3390/s100908526
Rao MR, Moon HS, Schenk TMH, Becker D, Mazarei M, Stewart CN Jr. FLP/FRT Recombination from Yeast: Application of a Two Gene Cassette Scheme as an Inducible System in Plants. Sensors. 2010; 10(9):8526-8535. https://doi.org/10.3390/s100908526
Chicago/Turabian StyleRao, Murali R., Hong S. Moon, Tobias M. H. Schenk, Dirk Becker, Mitra Mazarei, and C. Neal Stewart, Jr. 2010. "FLP/FRT Recombination from Yeast: Application of a Two Gene Cassette Scheme as an Inducible System in Plants" Sensors 10, no. 9: 8526-8535. https://doi.org/10.3390/s100908526