A Online NIR Sensor for the Pilot-Scale Extraction Process in Fructus Aurantii Coupled with Single and Ensemble Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
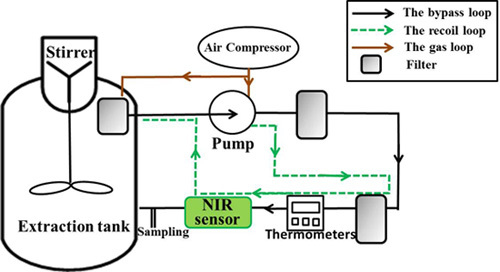
2.2. Preparation of Samples
| Extraction | Extraction Time | ||
|---|---|---|---|
| process | Heating | 0–1 h | 1–1.5 h |
| 1st extraction | 3 min | 4 min | 4 min |
| 2nd extraction | 5 min | 5 min | 5 min |
| 3rd extraction | 5 min | 6 min | 10 min |

2.3. NIR Equipment and Measurement
2.4. HPLC Method for Fructus aurantii
2.5. Preprocessing and Variable Selection Methods
2.6. Software and Data Analysis
3. Results and Discussion
3.1. NIR Spectrum Characteristics of Fructus aurantii

3.2. Quantitative analysis of Naringin, Hesperidin and Neohesperidin by HPLC Method

| Compound | Minimum Value (mg/mL) | Maximum Value (mg/mL) | Average Value (mg/mL) |
|---|---|---|---|
| Hesperidin | 0.0146 | 0.1889 | 0.0750 |
| Naringin | 0.2303 | 2.5504 | 1.0002 |
| Neohesperidin | 0.1893 | 2.0272 | 0.7814 |
3.3. NIR Optimum Result of Preprocessing Methods and Latent Factors

| Quality Parameter | Preprocessing Methods | Latent Factors | Calibration Set | Validation Set | Prediction Set | |||
|---|---|---|---|---|---|---|---|---|
| RMSEC * | R2 | RMSECV * | R2 | RMSEP * | R2 | |||
| Hesperidin | RAW | 5 | 0.0067 | 0.9760 | 0.0096 | 0.9530 | 0.0158 | 0.9237 |
| SG9 | 5 | 0.0073 | 0.9721 | 0.0096 | 0.9531 | 0.0159 | 0.9222 | |
| SG11 | 5 | 0.0074 | 0.9710 | 0.0096 | 0.9528 | 0.0160 | 0.9217 | |
| SG11 + 1D | 3 | 0.0155 | 0.8724 | 0.0274 | 0.6195 | 0.0349 | 0.6258 | |
| SG11 + 2D | 1 | 0.0339 | 0.3928 | 0.0496 | 0.2487 | 0.0665 | 0.3582 | |
| Normalize | 5 | 0.0059 | 0.9817 | 0.0095 | 0.9541 | 0.0156 | 0.9255 | |
| MSC | 5 | 0.0051 | 0.9863 | 0.0091 | 0.9583 | 0.0166 | 0.9119 | |
| SNV | 5 | 0.0052 | 0.9857 | 0.0090 | 0.9585 | 0.0160 | 0.9209 | |
| Naringin | RAW | 5 | 0.0892 | 0.9766 | 0.1334 | 0.9499 | 0.1551 | 0.9544 |
| SG9 | 5 | 0.0916 | 0.9753 | 0.1234 | 0.9569 | 0.1567 | 0.9535 | |
| SG11 | 5 | 0.0922 | 0.9750 | 0.1222 | 0.9578 | 0.1568 | 0.9534 | |
| SG11 + 1D | 3 | 0.2323 | 0.8411 | 0.4003 | 0.5469 | 0.4342 | 0.6424 | |
| SG11 + 2D | 1 | 0.4991 | 0.2665 | 0.6717 | 0.2757 | 0.8544 | 0.3845 | |
| Normalize | 5 | 0.0854 | 0.9785 | 0.1395 | 0.9450 | 0.1688 | 0.9460 | |
| MSC | 5 | 0.0721 | 0.9847 | 0.1347 | 0.9487 | 0.1738 | 0.9427 | |
| SNV | 5 | 0.0744 | 0.9837 | 0.1349 | 0.9486 | 0.1758 | 0.9414 | |
| Neohesperidin | RAW | 5 | 0.0774 | 0.9705 | 0.1108 | 0.9420 | 0.1197 | 0.9545 |
| SG9 | 5 | 0.0796 | 0.9688 | 0.1062 | 0.9467 | 0.1201 | 0.9542 | |
| SG11 | 5 | 0.0801 | 0.9684 | 0.1053 | 0.9476 | 0.1200 | 0.9543 | |
| SG11+1D | 3 | 0.1848 | 0.8318 | 0.3164 | 0.5264 | 0.3386 | 0.6356 | |
| SG11+2D | 1 | 0.3635 | 0.3495 | 0.4943 | 0.1556 | 0.6671 | 0.4145 | |
| Normalize | 5 | 0.0724 | 0.9742 | 0.1167 | 0.9356 | 0.1288 | 0.9472 | |
| MSC | 5 | 0.0604 | 0.9821 | 0.1116 | 0.9411 | 0.2240 | 0.8405 | |
| SNV | 5 | 0.0622 | 0.9810 | 0.1120 | 0.9407 | 0.1330 | 0.9438 | |
3.4. Different Variable Selection for PLS Models of Three Compounds in Fructus aurantii


3.5. Comparison of the Model Results by SiPLS and PLS Methods
| Quality Parameter | Model | Calibration Set | Validation Set | Prediction Set | ||||
|---|---|---|---|---|---|---|---|---|
| RMSEC * | R2 | RMSECV * | R2 | RMSEP * | R2 | |||
| Hesperidin | PLS | 0.0067 | 0.9760 | 0.0096 | 0.9530 | 0.0158 | 0.9237 | |
| SiPLS | 0.0066 | 0.9770 | 0.0079 | 0.9687 | 0.0155 | 0.9261 | ||
| Naringin | PLS | 0.0892 | 0.9776 | 0.1334 | 0.9499 | 0.1551 | 0.9544 | |
| SiPLS | 0.0469 | 0.9953 | 0.0550 | 0.9915 | 0.1493 | 0.9577 | ||
| Neohesperidin | PLS | 0.0774 | 0.9705 | 0.1108 | 0.9420 | 0.1197 | 0.9545 | |
| SiPLS | 0.0406 | 0.9919 | 0.0502 | 0.9881 | 0.1122 | 0.9599 | ||

3.6. Comparison of the Model Results by PLS Methods and Bagging-PLS



4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- U.S. Food Drug Administration. Guidance for Industry PAT: A Framework for Innovative Pharmaceutical Development Manufacturing and Quality Assurance. U.S. Food Drug Administration: Rockville, MD, USA, 2004. Available online: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRugulatoryInformation/Guidance/UCM070305.pdf (accessed on 1 March 2015). [Google Scholar]
- Killner, M.; Rohwedder, J.; Pasquini, C. A PLS regression model using NIR spectroscopy for online monitoring of the biodiesel production reaction. Fuel 2011, 90, 3268–3273. [Google Scholar] [CrossRef]
- Collell, C.; Gou, P.; Arnau, J.; Muñoz, I.; Comaposada, J. NIR technology for online determination of superficial aw and moisture content during the drying process of fermented sausages. Food Chem. 2012, 135, 1750–1755. [Google Scholar] [CrossRef] [PubMed]
- Marín-González, O.; Kuang, B.; Quraishi, M.; Munóz-García, M.; Mouazen, A. Online measurement of soil properties without direct spectral response in near infrared spectral range. Soil Till. Res. 2013, 132, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Sui, C.; Xu, B.; Ai, L.; Ma, Q.; Shi, X.; Qiao, Y. Multivariate detection limits of online NIR model for extraction process of chlorogenic acid from Lonicera japonica. J. Pharm. Biomed. Anal. 2013, 77, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Li, B.; Liu, X. Online determination for concentrating process of red ginseng alcohol extract using near-infrared spectroscopy. Chin. Pharm. J. 2006, 40, 1897–1899. [Google Scholar]
- Jin, Y.; Wu, Z.; Liu, X.; Wu, Y. Near infrared spectroscopy in combination with chemometrics as a process analytical technology (PAT) tool for online quantitative monitoring of alcohol precipitation. J. Pharm. Biomed. Anal. 2013, 77, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, G. Near-Infrared spectroscopy technology for online monitoring of the column separation and purification process of active components of Centella asiatica L. Urban. Spectrosc. Spect. Anal. 2013, 33, 98–101. [Google Scholar]
- Wan, W.; Jiang, H.; Shen., J.; Liu, Y.; Zhu, J.; Luo, H. Moisture and content of semi-manufactured gingko leaf dispersible tablets using near-infrared diffuse reflectance spectroscopy. Chin. J. Pharm. Anal. 2013, 33, 63–68. [Google Scholar]
- Geng, Z.; Hu, H.; Li, S.; Wang, M.; Mei, L.; Sun, X. Studies on NIR spectroscopy of Chuanhong Huoxue capsules extraction process. Chin. Pharm. 2012, 21, 14–15. [Google Scholar]
- Jiang, B.; Wang, Q.; Cai, R. Quantitative models for baicalin content using NIR technology for the study of Shang Jie Plaster. Spectrosc. Spect. Anal. 2013, 33, 74–77. [Google Scholar]
- Jin, Y.; Yang, K.; Chen, M.; Liu, X.; Chen, Y.; Long, C.; Wu, Y. In-line monitoring of blending process of Zhongsheng pill powder using miniature near-infrared analyzer. Chin. J. Chin. Mater. Med. 2011, 36, 2963–2966. [Google Scholar]
- Zhang, M.; Sheng, P.; Mu, Y.; Li, W.; Yu, H.; Harada, H.; Li, Y. Rapid and accurate determination of VFAs and ethanol in the effluent of an anaerobic H2-producing bioreactor using near-infrared spectroscopy. Water Res. 2009, 43, 1823–1830. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Xu, E.; Long, J.; Zhang, Y.; Wang, F.; Xu, X.; Jin, Z.; Jiao, A. Monitoring of fermentation process parameters of Chinese rice wine using attenuated total reflectance mid-infrared spectroscopy. Food Control 2015, 50, 405–412. [Google Scholar] [CrossRef]
- Mevik, B.; Segtnan, V.; Næs, T. Ensemble methods and partial least squares regression. J. Chemometr. 2004, 18, 498–507. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar]
- Viscarra Rossel, R. Robust modelling of soil diffuse reflectance spectra by bagging-partial least squares regression. J. Near Infrared Spectrosc. 2007, 15, 39–47. [Google Scholar] [CrossRef]
- National Commission of Chinese Pharmacopoeia. Pharmacopeia of People’s Republic of China; Chinese Medical Science and Technology Press: Beijing, China, 2010. [Google Scholar]
- Tong, L.; Zhou, D.; Gao, J.; Zhu, Y.; He, S.; Bi, K. Simultaneous determination of naringin, hesperidin, neohesperidin, naringenin and hesperetin of Fractus. aurantii extract in rat plasma by liquid chromatography tandem mass spectrometry. J. Pharm. Biomed. Anal. 2012, 58, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wang, H.; Xu, Y.; Dong, J.; Hashi, Y.; Chen, S. Identification of antioxidants in Fructus. aurantii and its quality evaluation using a new online combination of analytical techniques. Food Chem. 2012, 134, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Benavente-Garcia, O.; Castillo, J. Update on uses and properties of citrus flavonoids: New findings in anticancer, cardiovascular, and anti-inflammatory activity. J. Agric. Food Chem. 2008, 56, 6185–6205. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Garg, S.; Zaneveld, L.; Singla, A. Chemistry and pharmacology of the citrus bioflavonoid hesperidin. Phytother. Res. 2001, 15, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Huang, X.; Xing, Z.; Qiu, X.; Wang, Y.; Fan, R.; Liu, W.; Ren, P.; Liu, Z.; Zhou, H. Meranzin hydrate induces similar effect to Fructus Aurantii. on intestinal motility through activation of H1 histamine receptors. J. Gastrointest. Surg. 2011, 15, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Sui, C.; Wu, Z.; Peng, Y.; Zou, L.; Pei, Y.; Shi, X.; Qiao, Y. Validation of NIR Model for Online Monitoring of Flos Lonicera Japonica Extraction Process with Different Batches of Materials. Int. J. Online Eng. 2013, 9, 44–48. [Google Scholar] [CrossRef]
- Rinnan, Å.; van den Berg, F.; Engelsen, S. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Roggo, Y.; Chalus, P.; Maurer, L.; Lema-Martinez, C.; Edmond, A.; Jent, N. A review of near infrared spectroscopy and chemometrics in pharmaceutical technologies. J. Pharm. Biomed. Anal. 2007, 44, 683–700. [Google Scholar] [CrossRef] [PubMed]
- Barnes, R.; Dhanoa, M.; Lister, J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Isaksson, T.; Naes, T. The effect of multiplicative scatter correction (MSC) and linearity improvement in NIR spectroscopy. Appl. Spectrosc. 1988, 42, 1273–1284. [Google Scholar] [CrossRef]
- Luypaert, J.; Massart, D.; Heyden, Y. Near-infrared spectroscopy applications in pharmaceutical analysis. Talanta 2007, 72, 865–883. [Google Scholar] [CrossRef] [PubMed]
- Norgaard, L.; Saudland, A.; Wagner, J.; Nielsen, J.; Munck, L.; Engelsen, S. Interval partial least-squares regression (iPLS): A comparative chemometric study with an example from near-infrared spectroscopy. Appl. Spectrosc. 2000, 54, 413–419. [Google Scholar] [CrossRef]
- Chen, H.; Pan, T.; Chen, J.; Lu, Q. Waveband selection for NIR spectroscopy analysis of soil organic matter based on SG smoothing and MWPLS methods. Chemometr. Intell. Lab. Syst. 2011, 107, 139–146. [Google Scholar] [CrossRef]
- Quality & Technology. Available online: http://www.models.kvl.dk/ (accessed on 12 December 2014).
- Yamatera, H.; Fitzpatrick, B.; Gordok, G. Near infrared spectra of water and aqueous solution. Mol. Struct. 1964, 14, 268–278. [Google Scholar]
- Patricia, P.; Sanchez, M.; Dolores, P.; Guerrero, J.; Ana, G. Evaluating NIR instruments for quantitative and qualitative assessment of intact apple quality. Sci. Food Aric. 2009, 89, 781–790. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, X.; Li, Y.; Wu, Z.; Zhang, Q.; Zheng, Z.; Shi, X.; Qiao, Y. A Online NIR Sensor for the Pilot-Scale Extraction Process in Fructus Aurantii Coupled with Single and Ensemble Methods. Sensors 2015, 15, 8749-8763. https://doi.org/10.3390/s150408749
Pan X, Li Y, Wu Z, Zhang Q, Zheng Z, Shi X, Qiao Y. A Online NIR Sensor for the Pilot-Scale Extraction Process in Fructus Aurantii Coupled with Single and Ensemble Methods. Sensors. 2015; 15(4):8749-8763. https://doi.org/10.3390/s150408749
Chicago/Turabian StylePan, Xiaoning, Yang Li, Zhisheng Wu, Qiao Zhang, Zhou Zheng, Xinyuan Shi, and Yanjiang Qiao. 2015. "A Online NIR Sensor for the Pilot-Scale Extraction Process in Fructus Aurantii Coupled with Single and Ensemble Methods" Sensors 15, no. 4: 8749-8763. https://doi.org/10.3390/s150408749