A Homogenous Fluorescence Quenching Based Assay for Specific and Sensitive Detection of Influenza Virus A Hemagglutinin Antigen
Abstract
:1. Introduction

2. Experimental Section
2.1. Reagents
2.2. Preparation of Antibody-Labeled Au NPs
2.3. Preparation of Glycan-Functionalized Quantum Dots
2.4. Characterization of Nanoparticles
2.5. Detection of AIV HA Antigen by Homogeneous Fret Assay
3. Results and Discussion
3.1. Preparation of Gly-QDs and Ab-Au NPs

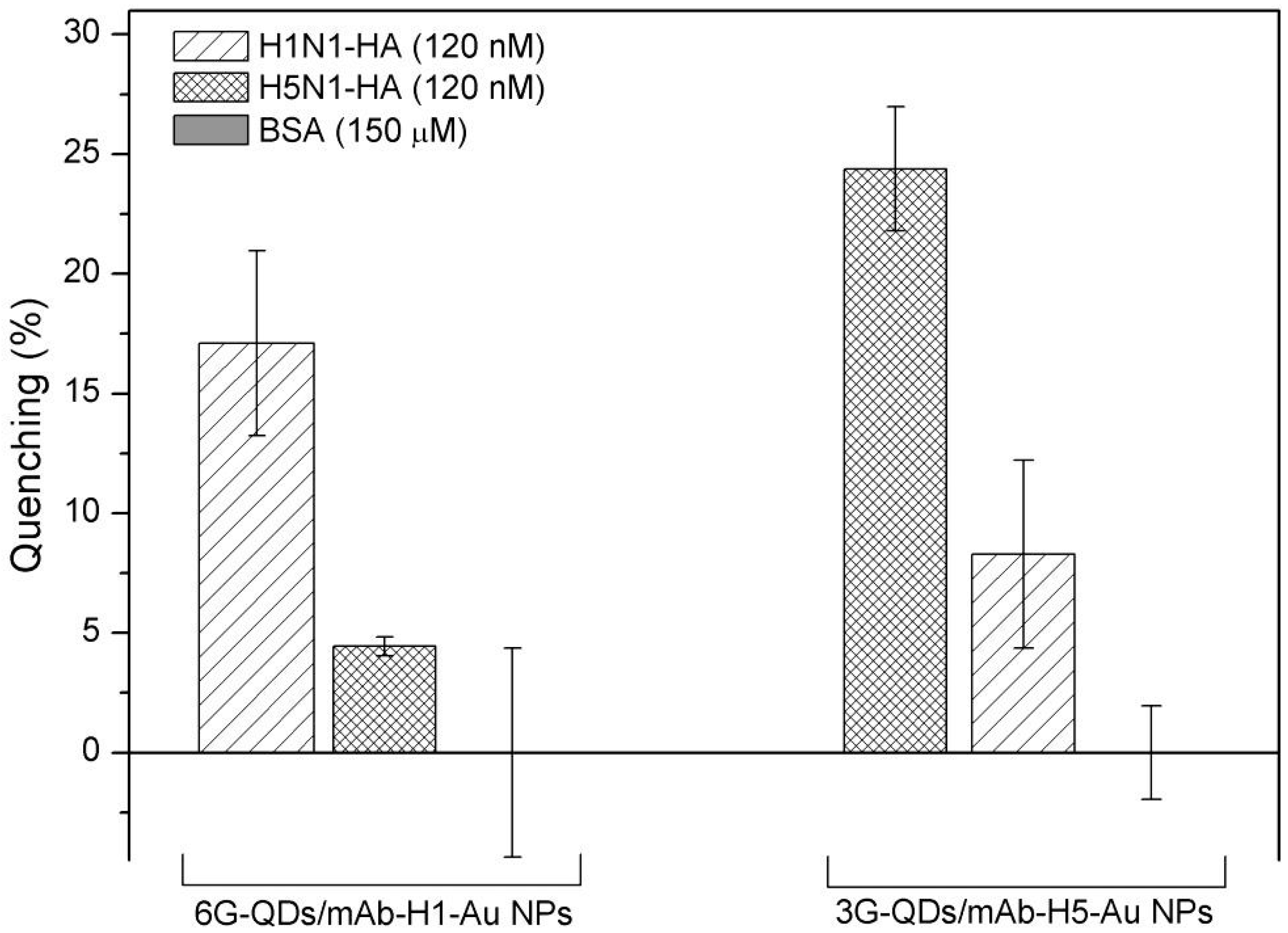
3.2. Detection of Influenza Virus HAs

| Sensor | Probe | HA Derived Influenza Type | Sensitivity or Affinity | Remark | Reference |
|---|---|---|---|---|---|
| SPR | Glycan-ligands | H1N1 | Kd 1.5 µM (10 µg/mL, ~17 µM) | Low affinity and non-specific interaction. | [17] |
| QCM | Glycan | H5N3 | Kd 14.4 nM | Good affinity. | [42] |
| SPR | Glycan | H5N1 | Kd 1.6 nM | High affinity; specific substrate (Biacore chip) required. | [43] |
| Waveguide | Antibody | H1N1, H5N1 | 1 nM | Dissociation of dye from antibody may decrease sensitivity. | [44] |
| SPR | DNA Aptamer | H5N1 | Kd 4.65 nM | Good affinity. | [45] |
| Electrochemical | Immuniliposome-Ru | H1N1 | 3 × 10−14 g/mL (12 fM) | Very high sensitivity; complex probe preparation step. | [46] |
| SPR | RNA Aptamer | H3N2 | Kd 120 pM | High affinity; unstable RNA aptamer probe. | [47] |
| Interferometry | RNA Aptamer | H3N2 | 10 nM | RNA aptamer; relative low sensitivity. | [48] |
| SPR | RNA Aptamer | H1N1 | Kd 67 fM | High affinity. | [49] |
| Field Effect Transistor (FET) | Glycan | H5N1, H1N1 | 50 aM | Very high sensitivity. | [50] |

4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- WHO (2014). Available online: http://www.who.int/mediacentre/factsheets/fs211/en/ (accessed on 18 July 2014).
- Peiris, J.S.M.; de Jong, M.D.; Guan, Y. Avian influenza virus (H5N1): A threat to human health. Clin. Microbiol. Rev. 2007, 20, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef] [PubMed]
- Reid, A.H.; Taubenberger, J.K.; Fanning, T.G. The 1918 Spanish influenza: Integrating history and biology. Microb. Infect. 2001, 3, 81–87. [Google Scholar] [CrossRef]
- Stevens, J.; Blixt, O.; Glaser, L.; Taubenberger, J.K.; Palese, P.; Paulson, J.C.; Wilson, I.A. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol. 2006, 355, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, R.S.; Pfeiffer, D.; Otte, J. Evaluating the control of HPAIV H5N1 in Vietnam: Virus transmission within infected flocks reported before and after vaccination. BMC Vet. Res. 2010, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Amano, Y.; Cheng, Q. Detection of influenza virus: Traditional approaches and development of biosensors. Anal. Bioanal. Chem. 2005, 381, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.S.; Zambon, M.C. Molecular diagnosis of influenza. Rev. Med. Virol. 2002, 12, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Charlton, B.; Crossley, B.; Hietala, S. Conventional and future diagnostics for avian influenza. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Zhou, E.M.; Chan, M.; Heckert, R.A.; Riva, J.; Cantin, M.F. Evaluation of a competitive ELISA for detection of antibodies against avian influenza virus nucleoprotein. Avian Dis. 1998, 42, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Rowe, T.; Abernathy, R.A.; Hu-Primmer, J.; Thompson, W.W.; Lu, X.; Lim, W.; Fukuda, K.; Cox, N.J.; Katz, J.M. Detection of antibody to avian influenza a (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 1999, 37, 937–943. [Google Scholar] [PubMed]
- WHO Global Influenza Surveillance Network. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza; WHO Press: Geneva, Switzerland, 2011. [Google Scholar]
- Rogers, G.N.; Paulson, J.C. Receptor determinants of human and animal influenza virus isolates: Differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 1983, 127, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Couceiro, J.N.S.S.; Paulson, J.C.; Baum, L.G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993, 29, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.N.; D'Souza, B.L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 1989, 173, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Critchley, P.; Dimmock, N.J. Binding of an influenza A virus to a neomembrane measured by surface plasmon resonance. Bioorg. Med. Chem. 2004, 12, 2773–2780. [Google Scholar] [CrossRef] [PubMed]
- Mandenius, C.-F.; Wang, R.; Aldén, A.; Bergström, G.; Thébault, S.; Lutsch, C.; Ohlson, S. Monitoring of influenza virus hemagglutinin in process samples using weak affinity ligands and surface plasmon resonance. Anal. Chim. Acta 2008, 623, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, S.; Hinou, H.; Ebihara, D.; Sato, R.; Kuroiwa, S.; Nakanishi, T.; Nishimura, S.-I.; Osaka, T. Attomolar detection of influenza a virus hemagglutinin human h1 and avian H5 using glycan-blotted field effect transistor biosensor. Anal. Chem. 2013, 85, 5641–5644. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.B.; Awazu, K.; Fujimaki, M.; Shimizu, K. Neu5Acα2,6Gal and Neu5Acα2,3Gal receptor specificities on influenza viruses determined by a waveguide-mode sensor. Acta Biomater. 2013, 9, 5080–5087. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Gaston, M.A.; Weiss, A.A.; Zhang, P. Colorimetric viral detection based on sialic acid stabilized gold nanoparticles. Biosens. Bioelectron. 2013, 42, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Roda, A.; Guardigli, M.; Michelini, E.; Mirasoli, M. Nanobioanalytical luminescence: Förster-type energy transfer methods. Anal. Bioanal. Chem. 2009, 393, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-C.; Huang, Y.-H. Nucleic acid sandwich hybridization assay with quantum dot-induced fluorescence resonance energy transfer for pathogen detection. Sensors 2012, 12, 16660–16672. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Zhao, H.; Liu, M.; Chen, Y.; Quan, X. Detection of influenza A virus based on fluorescence resonance energy transfer from quantum dots to carbon nanotubes. Anal. Chim. Acta 2012, 723, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Choi, Y.; Kim, J.; Park, E.; Song, R. Positively charged compact quantum dot–DNA complexes for detection of nucleic acids. Chem. Phys. Chem. 2009, 10, 806–811. [Google Scholar] [PubMed]
- Sebestyén, Z.; Nagy, P.; Horváth, G.; Vámosi, G.; Debets, R.; Gratama, J.W.; Alexander, D.R.; Szöllősi, J. Long wavelength fluorophores and cell-by-cell correction for autofluorescence significantly improves the accuracy of flow cytometric energy transfer measurements on a dual-laser benchtop flow cytometer. Cytometry 2002, 48, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Medintz, I.L.; Clapp, A.R.; Mattoussi, H.; Goldman, E.R.; Fisher, B.; Mauro, J.M. Self-assembled nanoscale biosensors based on quantum dot FRET donors. Nat. Mater. 2003, 2, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Clapp, A.R.; Medintz, I.L.; Mauro, J.M.; Fisher, B.R.; Bawendi, M.G.; Mattoussi, H. Fluorescence resonance energy transfer between quantum dot donors and dye-labeled protein acceptors. J. Am. Chem. Soc. 2003, 126, 301–310. [Google Scholar] [CrossRef]
- Algar, W.R.; Krull, U. Quantum dots as donors in fluorescence resonance energy transfer for the bioanalysis of nucleic acids, proteins, and other biological molecules. Anal. Bioanal. Chem. 2008, 391, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Sapsford, K.E.; Berti, L.; Medintz, I.L. Materials for fluorescence resonance energy transfer analysis: beyond traditional donor–acceptor combinations. Angew. Chem. Int. Ed. 2006, 45, 4562–4589. [Google Scholar] [CrossRef]
- Jain, P.K.; El-Sayed, I.H.; El-Sayed, M.A. Au nanoparticles target cancer. Nano Today 2007, 2, 18–29. [Google Scholar] [CrossRef]
- Tang, B.; Cao, L.; Xu, K.; Zhuo, L.; Ge, J.; Li, Q.; Yu, L. A new nanobiosensor for glucose with high sensitivity and selectivity in serum based on fluorescence resonance energy transfer (FRET) between CdTe quantum dots and Au nanoparticles. Chem. Eur. J. 2008, 14, 3637–3644. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Wang, X.; Duan, L.; Gao, W.; Ji, L.; Tang, B. A new nanoprobe based on FRET between functional quantum dots and gold nanoparticles for fluoride anion and its applications for biological imaging. Biosens. Bioelectron. 2012, 36, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Wang, C.; Pan, X.; Liu, S. A high-throughput homogeneous immunoassay based on Förster resonance energy transfer between quantum dots and gold nanoparticles. Anal. Chim. Acta 2013, 763, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, Y.; Luo, Y.; Shen, F.; Sun, C. Efficient fluorescence resonance energy transfer between oppositely charged CdTe quantum dots and gold nanoparticles for turn-on fluorescence detection of glyphosate. Talanta 2014, 125, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Frens, G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nature 1973, 241, 20–22. [Google Scholar]
- Hermanson, G.T.; Hermanson, G.T. Chapter 24—Preparation of Colloidal Gold-Labeled Proteins, Bioconjugate Techniques, 2nd ed.; Academic Press: New York, NY, USA, 2007; pp. 924–935. [Google Scholar]
- Samanta, A.; Zhou, Y.; Zou, S.; Yan, H.; Liu, Y. Fluorescence quenching of quantum dots by gold nanoparticles: a potential long range spectroscopic ruler. Nano Lett. 2014, 14, 5052–5057. [Google Scholar] [CrossRef] [PubMed]
- Springsteen, G.; Wang, B. A detailed examination of boronic acid-diol complexation. Tetrahedron 2002, 58, 5291–5300. [Google Scholar] [CrossRef]
- Ferrier, R.J.; Prasad, D. 1360. Boric acid derivatives as reagents in carbohydrate chemistry. Part VI. Phenylboronic acid as a protecting group in disaccharide synthesis. J. Chem. Soc. 1965, 7429–7432. [Google Scholar] [CrossRef]
- Brewer, S.H.; Allen, A.M.; Lappi, S.E.; Chasse, T.L.; Briggman, K.A.; Gorman, C.B.; Franzen, S. Infrared Detection of a phenylboronic acid terminated alkane thiol monolayer on gold surface. Langmuir 2004, 20, 5512–5520. [Google Scholar] [CrossRef] [PubMed]
- Dulkeith, E.; Morteani, A.C.; Niedereichholz, T.; Klar, T.A.; Feldmann, J.; Levi, S.A.; van Veggel, F.C.J.M.; Reinhoudt, D.N.; Möller, M.; Gittins, D.I. Fluorescence quenching of dye molecules near gold nanoparticles: radiative and nonradiative effects. Phys. Rev. Lett. 2002, 89. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Kawagishi, S.; Masuda, M.; Suzuki, T. Binding kinetics of sulfatide with influenza A virus hemagglutinin. Glycoconjugate J. 2013, 30, 709–716. [Google Scholar] [CrossRef]
- Suenaga, E.; Mizuno, H.; Penmetcha, K.K.R. Monitoring influenza hemagglutinin and glycan interactions using surface plasmon resonance. Biosens. Bioelectron. 2012, 32, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.B.; Awazu, K.; Fujimaki, M. Detection of influenza viruses by a waveguide-mode sensor. Anal. Methods 2010, 2, 1880–1884. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, J.; Jiang, T.; Kwon, Y.M.; Lu, H.; Jiao, P.; Liao, M.; Li, Y. Selection and characterization of DNA aptamers for use in detection of avian influenza virus H5N1. J. Virol. Methods 2013, 189, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Egashira, N.; Morita, S.-I.; Hifumi, E.; Mitoma, Y.; Uda, T. Attomole detection of hemagglutinin molecule of influenza virus by combining an electrochemiluminescence sensor with an immunoliposome that encapsulates a ru complex. Anal. Chem. 2008, 80, 4020–4025. [Google Scholar] [CrossRef] [PubMed]
- Misono, T.S.; Kumar, P.K. Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal. Biochem. 2005, 342, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Awazu, K.; Tominaga, J.; Kumar, P.K. Monitoring biomolecular interactions on a digital versatile disk: A bioDVD platform technology. ACS Nano 2008, 2, 1885–1895. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Kumar, P.K. Aptamers that bind to the hemagglutinin of the recent pandemic influenza virus H1N1 and efficiently inhibit agglutination. Acta Biomater. 2013, 9, 8932–8941. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, S.; Kuroiwa, S.; Kimura, M.; Cheng, S.; Osaka, T. Effect of the size of receptor in allergy detection using field effect transistor biosensor. Electrochimi. Acta 2013, 110, 146–151. [Google Scholar] [CrossRef]
- Steward, M.W.; Lew, A.M. The importance of antibody affinity in the performance of immunoassays for antibody. J. Immunol. Methods 1985, 78, 173–190. [Google Scholar] [CrossRef] [PubMed]
- Amiry-Moghaddam, M.; Ottersen, O.P. Immunogold cytochemistry in neuroscience. Nat. Neurosci. 2013, 16, 798–804. [Google Scholar] [CrossRef] [PubMed]
- Beebe, D.J.; Mensing, G.A.; Walker, G.M. Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 2002, 4, 261–286. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Neethirajan, S. A Homogenous Fluorescence Quenching Based Assay for Specific and Sensitive Detection of Influenza Virus A Hemagglutinin Antigen. Sensors 2015, 15, 8852-8865. https://doi.org/10.3390/s150408852
Chen L, Neethirajan S. A Homogenous Fluorescence Quenching Based Assay for Specific and Sensitive Detection of Influenza Virus A Hemagglutinin Antigen. Sensors. 2015; 15(4):8852-8865. https://doi.org/10.3390/s150408852
Chicago/Turabian StyleChen, Longyan, and Suresh Neethirajan. 2015. "A Homogenous Fluorescence Quenching Based Assay for Specific and Sensitive Detection of Influenza Virus A Hemagglutinin Antigen" Sensors 15, no. 4: 8852-8865. https://doi.org/10.3390/s150408852






