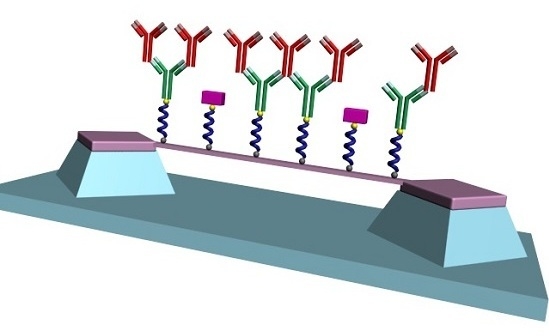
Diazonium Chemistry for the Bio-Functionalization of Glassy Nanostring Resonator Arrays
Abstract
:1. Introduction
2. Experimental
2.1. Fabrication of SiCN Nanostrings

2.2. Laser Interferometry Measurement
2.3. Synthesis of Aryl Diazonium Salt
2.4. Diazonium Induced SiCN Surface Modification
2.5. XPS
2.6. Resonator Surface Biofunctionalization

2.6.1. Resonator Surface Biofunctionalization
2.6.2. Activation of Carboxyl Groups
2.6.3. Immobilization of Recognition Bioreceptor
2.6.4. Capture of Target and Negative Control
2.7. Helium Ion Microscopy of Functionalized Nanoresonators
3. Results and Discussion
3.1. SiCN Nanomechanical Resonator Array

3.2. One-Step Modification of SiCN Surface

3.3. Covalent Immobilization of Anti-Rabbit IgG to SiCN Resonator

| Nanostring Width (nm) | 300 | 250 | 200 | 180 |
|---|---|---|---|---|
| Average unloaded frequency f0 (MHz) | 14.542 ± 0.0080 | 14.447 ± 0.0087 | 14.4135 ± 0.0157 | 14.4232 ± 0.0184 |
| Average shift frequency due to probe Δf (kHz) | 250 ± 23 | 265 ± 21 | 320 ± 20 | 420 ± 31 |
| Added mass of probe Δm (fg) | 17 ± 1.56 | 15 ± 1.2 | 14.7 ± 0.92 | 17.3 ± 1.3 |
| Added mass-per-area of probe Δm/s (fg/µm2) | 1.6 ± 0.15 | 1 ± 0.13 | 1.95 ± 0.12 | 2.50 ± 0.19 |
3.4. Specific Detection of Target Rabbit IgG Attached to Resonator
| Resonator String Width (nm) | 300 | 250 | 200 | 180 |
|---|---|---|---|---|
| Average unloaded frequency f0 (MHz) | 14.411 ± 0.0089 | 14.4298 ± 0.0175 | 14.5155 ± 0.0216 | 14.6142 ± 0.0147 |
| Average shift frequency due to probe and target Δf (kHz) | 395 ± 51 | 470 ± 46 | 541 ± 37 | 621 ± 16 |
| Added mass of probe and target Δm (fg) | 27 ± 3.5 | 27 ± 2.6 | 24.5 ± 1.5 | 25.3 ± 0.6 |
| Added mass-per-area of probe and target Δm/s (fg/µm2) | 2.6 ± 0.3 | 3.0 ± 0.3 | 3.3 ± 0.22 | 3.66 ± 0.09 |
| Added mass-per-area of target Δm/s (fg/µm2) | 0.96 ± 0.29 | 1.29 ± 0.30 | 1.42 ± 0.23 | 1.07 ± 0.21 |
| Total number of target molecules | 40,500 ± 1200 | 46,500 ± 1100 | 43,000 ± 6800 | 30,000 ± 5800 |
| Resonator String Width (nm) | 300 | 250 | 200 | 180 |
|---|---|---|---|---|
| Average unloaded frequency (MHz) | 14.31 ± 0.035 | 14.20 ± 0.04 | 14.0234 ± 0.0140 | 13.97 ± 0.08 |
| Average shift frequency due to probe and control (kHz) | 267 ± 19 | 295 ± 18 | 345.0 ± 28 | 440 ± 31 |
| Average shift frequency due to control (kHz) | 15 ± 33 | 27 ± 24 | 23 ± 47 | 24 ± 50 |
| Average shift frequency ratio of control to target (percentage) | 10.6% | 13.3% | 9.7% | 12.8% |
3.5. HIM Protein Observation on the Resonator Surface

4. Conclusions/Outlook
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Haes, A.J.; Chang, L.; Klein, W.L.; van Duyne, R.P. Detection of a biomarker for alzheimer’s disease from synthetic and clinical samples using a nanoscale optical biosensor. J. Am. Chem. Soc. 2005, 127, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Tothill, I.E. Biosensors for cancer markers diagnosis. Semin. Cell Dev. Boil. 2009, 20, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, K.A.; Taylor, D. Fluorescent-protein biosensors: New tools for drug discovery. Trends Biotechnol. 1998, 16, 135–140. [Google Scholar] [CrossRef]
- Keusgen, M. Biosensors: New approaches in drug discovery. Naturwissenschaften 2002, 89, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A. Optical biosensors in drug discovery. Nat. Rev. Drug Discov. 2002, 1, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.C.; Mastali, M.; Gau, V.; Suchard, M.A.; Møller, A.K.; Bruckner, D.A.; Babbitt, J.T.; Li, Y.; Gornbein, J.; Landaw, E.M. Use of electrochemical DNA biosensors for rapid molecular identification of uropathogens in clinical urine specimens. J. Clin. Microbial. 2006, 44, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Gartia, M.R.; Braunschweig, B.; Chang, T.-W.; Moinzadeh, P.; Minsker, B.S.; Agha, G.; Wieckowski, A.; Keefer, L.L.; Liu, G.L. The microelectronic wireless nitrate sensor network for environmental water monitoring. J. Environ. Monit. 2012, 14, 3068–3075. [Google Scholar] [CrossRef] [PubMed]
- Lieberzeit, P.A.; Dickert, F.L. Rapid bioanalysis with chemical sensors: Novel strategies for devices and artificial recognition membranes. Anal. Bioanal. Chem. 2008, 391, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- So, H.-M.; Won, K.; Kim, Y.H.; Kim, B.-K.; Ryu, B.H.; Na, P.S.; Kim, H.; Lee, J.-O. Single-walled carbon nanotube biosensors using aptamers as molecular recognition elements. J. Am. Chem. Soc. 2005, 127, 11906–11907. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Lee, B.; Liu, G.L.; Agarwal, A.; Lee, L.P. Label-free and highly sensitive biomolecular detection using sers and electrokinetic preconcentration. Lab Chip 2009, 9, 3360–3363. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, G.L.; Kim, J.; Mejia, Y.X.; Lee, L.P. Nanophotonic crescent moon structures with sharp edge for ultrasensitive biomolecular detection by local electromagnetic field enhancement effect. Nano Lett. 2005, 5, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Densmore, A.; Vachon, M.; Xu, D.X.; Janz, S.; Ma, R.; Li, Y.H.; Lopinski, G.; Delâge, A.; Lapointe, J.; Luebbert, C.C.; et al. Silicon photonic wire biosensor array for multiplexed real-time and label-free molecular detection. Opt. Lett. 2009, 34, 3598–3600. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Mehta, G.; Srivastava, S. Label-free detection techniques for protein microarrays: Prospects, merits and challenges. Proteomics 2010, 10, 731–748. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.X.; Densmore, A.; Delâge, A.; Waldron, P.; McKinnon, R.; Janz, S.; Lapointe, J.; Lopinski, G.; Mischki, T.; Post, E.; et al. Folded cavity soi microring sensors for highsensitivity and real time measurement ofbiomolecular binding. Opt. Express 2008, 16, 15137–15148. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.X.; Vachon, M.; Densmore, A.; Ma, R.; Delâge, A.; Janz, S.; Lapointe, J.; Li, Y.; Lopinski, G.; Zhang, D.; et al. Label-free biosensor array based on silicon-on-insulator ring resonators addressed using a wdm approach. Opt. Lett. 2010, 35, 2771–2773. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.L.; Lee, L.P. Nanowell surface enhanced raman scattering arrays fabricated by soft-lithography for label-free biomolecular detections in integrated microfluidics. Appl. Phys. Lett. 2005, 87. [Google Scholar] [CrossRef]
- Furuki, M.; Kameoka, J.; Craighead, H.G.; Isaacson, M.S. Surface plasmon resonance sensors utilizing microfabricated channels. Sens. Actuators B Chem. 2001, 79, 63–69. [Google Scholar] [CrossRef]
- Cui, Y.; Wei, Q.Q.; Park, H.K.; Lieber, C.M. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science 2001, 293, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Waggoner, P.S.; Craighead, H.G. Micro-and nanomechanical sensors for environmental, chemical, and biological detection. Lab Chip 2007, 7, 1238–1255. [Google Scholar] [CrossRef] [PubMed]
- Ilic, B.; Czaplewski, D.; Craighead, H.G.; Neuzil, P.; Campagnolo, C.; Batt, C. Mechanical resonant immunospecific biological detector. Appl. Phys. Lett. 2000, 77, 450–452. [Google Scholar] [CrossRef]
- Lavrik, N.V.; Sepaniak, M.J.; Datskos, P.G. Cantilever transducers as a platform for chemical and biological sensors. Rev. Sci. Instrum. 2004, 75, 2229–2253. [Google Scholar] [CrossRef]
- Baller, M.K.; Lang, H.P.; Fritz, J.; Gerber, C.; Gimzewski, J.K.; Drechsler, U.; Rothuizen, H.; Despont, M.; Vettiger, P.; Battiston, F.M.; et al. A cantilever array-based artificial nose. Ultramicroscopy 2000, 82, 1–9. [Google Scholar] [CrossRef]
- Raiteri, R.; Grattarola, M.; Butt, H.-J.; Skládal, P. Micromechanical cantilever-based biosensors. Sens. Actuators B Chem. 2001, 79, 115–126. [Google Scholar] [CrossRef]
- Villarroya, M.; Verd, J.; Teva, J.; Abadal, G.; Forsen, E.; Murano, F.P.; Uranga, A.; Figueras, E.; Montserrat, J.; Esteve, J.; et al. System on chip mass sensor based on polysilicon cantilevers arrays for multiple detection. Sens. Actuators A Phys. 2006, 132, 154–164. [Google Scholar] [CrossRef]
- Evoy, S.; Olkhovets, A.; Sekaric, L.; Parpia, J.M.; Craighead, H.G.; Carr, D.W. Temperature-dependent internal friction in silicon nanoelectromechanical systems. Appl. Phys. Lett. 2000, 77, 2397–2399. [Google Scholar] [CrossRef]
- Mohammad, M.; Dew, S.; Evoy, S.; Stepanova, M. Fabrication of sub-10nm silicon carbon nitride resonators using a hydrogen silsesquioxane mask patterned by electron beam lithography. Microelectron. Eng. 2011, 88, 2338–2341. [Google Scholar] [CrossRef]
- Fischer, L.M.; Wright, V.A.; Guthy, C.; Yang, N.; McDermott, M.T.; Buriak, J.M.; Evoy, S. Specific detection of proteins using nanomechanical resonators. Sens. Actuators B Chem. 2008, 134, 613–617. [Google Scholar] [CrossRef]
- Guthy, C.; Belov, M.; Janzen, A.; Quitoriano, N.J.; Singh, A.; Wright, V.A.; Finley, E.; Kamins, T.I.; Evoy, S. Large-scale arrays of nanomechanical sensors for biomolecular fingerprinting. Sens. Actuators B Chem. 2013, 187, 111–117. [Google Scholar] [CrossRef]
- Jensen, K.; Kim, K.; Zettl, A. An atomic-resolution nanomechanical mass sensor. Nat. Nanotechnol. 2008, 3, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Davis, Z.J.; Abadal, G.; Helbo, B.; Hansen, O.; Campabadal, F.; Pérez-Murano, F.; Esteve, J.; Figueras, E.; Verd, J.; Barniol, N. Monolithic integration of mass sensing nano-cantilevers with cmos circuitry. Sens. Actuators A Phys. 2003, 105, 311–319. [Google Scholar] [CrossRef]
- Yang, Y.; Callegari, C.; Feng, X.; Ekinci, K.; Roukes, M. Zeptogram-scale nanomechanical mass sensing. Nano Lett. 2006, 6, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Gil-Santos, E.; Ramos, D.; Martinez, J.; Fernandez-Regulez, M.; Garcia, R.; San Paulo, A.; Calleja, M.; Tamayo, J. Nanomechanical mass sensing and stiffness spectrometry based on two-dimensional vibrations of resonant nanowires. Nat. Nanotechnol. 2010, 5, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, J.; Kosaka, P.M.; Ruz, J.J.; San Paulo, A.; Calleja, M. Biosensors based on nanomechanical systems. Chem. Soc. Rev. 2013, 42, 1287–1311. [Google Scholar] [CrossRef] [PubMed]
- McKendry, R.; Zhang, J.; Arntz, Y.; Strunz, T.; Hegner, M.; Lang, H.P.; Baller, M.K.; Certa, U.; Meyer, E.; Güntherodt, H.-J. Multiple label-free biodetection and quantitative DNA-binding assays on a nanomechanical cantilever array. Proc. Natl. Acad. Sci. USA 2002, 99, 9783–9788. [Google Scholar] [CrossRef] [PubMed]
- Sadek, A.S.; Karabalin, R.B.; Du, J.; Roukes, M.L.; Koch, C.; Masmanidis, S.C. Wiring nanoscale biosensors with piezoelectric nanomechanical resonators. Nano Lett. 2010, 10, 1769–1773. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Patolsky, F.; Cui, Y.; Wang, W.U.; Lieber, C.M. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat. Biotechnol. 2005, 23, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Grieshaber, D.; MacKenzie, R.; Voros, J.; Reimhult, E. Electrochemical biosensors—Sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef] [Green Version]
- Göpel, W.; Heiduschka, P. Interface analysis in biosensor design. Biosens. Bioelectron. 1995, 10, 853–883. [Google Scholar] [CrossRef]
- Credou, J.; Volland, H.; Dano, J.; Berthelot, T. A one-step and biocompatible cellulose functionalization for covalent antibody immobilization on immunoassay membranes. J. Mater. Chem. B 2013, 1, 3277–3286. [Google Scholar] [CrossRef]
- Mévellec, V.; Roussel, S.; Tessier, L.; Chancolon, J.; Mayne-LʼHermite, M.; Deniau, G.; Viel, P.; Palacin, S. Grafting polymers on surfaces: A new powerful and versatile diazonium salt-based one-step process in aqueous media. Chem. Mater. 2007, 19, 6323–6330. [Google Scholar] [CrossRef]
- Mesnage, A.; Esnouf, S.; Jégou, P.; Deniau, G.; Palacin, S. Understanding the redox-induced polymer grafting process: A dual surface-solution analysis. Chem. Mater. 2010, 22, 6229–6239. [Google Scholar] [CrossRef]
- Pinson, J. Attachment of organic layers to materials surfaces by reduction of diazonium salts. In Aryl Diazonium. Salts: New Coupling Agents and Surface Science; Mohamed, M.C., Ed.; Wiley: Hoboken, NJ, USA, 2012. [Google Scholar]
- Ru, J.; Szeto, B.; Bonifas, A.; McCreery, R.L. Microfabrication and integration of diazonium-based aromatic molecular junctions. ACS Appl. Mater. Interfaces 2010, 2, 3693–3701. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.M.; Bergren, A.J.; McCreery, R.L. Derivatization of optically transparent materials with diazonium reagents for spectroscopy of buried interfaces. Anal. Chem. 2009, 81, 6972–6980. [Google Scholar] [CrossRef] [PubMed]
- Hurley, B.L.; McCreery, R.L. Covalent bonding of organic molecules to cu and al alloy 2024 T3 surfaces via diazonium ion reduction. J. Electrochem. Soc. 2004, 151, B252–B259. [Google Scholar] [CrossRef]
- Matrab, T.; Nguyen, M.N.; Mahouche, S.; Lang, P.; Badre, C.; Turmine, M.; Girard, G.; Bai, J.; Chehimi, M.M. Aryl diazonium salts for carbon fiber surface-initiated atom transfer radical polymerization. J. Adhes. 2008, 84, 684–701. [Google Scholar] [CrossRef]
- Shewchuk, D.M.; McDermott, M.T. Comparison of diazonium salt derived and thiol derived nitrobenzene layers on gold. Langmuir 2009, 25, 4556–4563. [Google Scholar] [CrossRef] [PubMed]
- Laurentius, L.; Stoyanov, S.R.; Gusarov, S.; Kovalenko, A.; Du, R.; Lopinski, G.P.; McDermott, M.T. Diazonium-derived aryl films on gold nanoparticles: Evidence for a carbon-gold covalent bond. ACS Nano 2011, 5, 4219–4227. [Google Scholar] [CrossRef] [PubMed]
- Kariuki, J.K.; McDermott, M.T. Formation of multilayers on glassy carbon electrodes via the reduction of diazonium salts. Langmuir 2001, 17, 5947–5951. [Google Scholar] [CrossRef]
- Mohammad, M.A.; Guthy, C.; Evoy, S.; Dew, S.K.; Stepanova, M. Nanomachining and clamping point optimization of silicon carbon nitride resonators using low voltage electron beam lithography and cold development. J. Vac. Sci. Technol. B 2010, 28, C6p36–C6p41. [Google Scholar] [CrossRef]
- Guthy, C.; Das, R.M.; Drobot, B.; Evoy, S. Resonant characteristics of ultranarrow sicn nanomechanical resonators. J. Appl. Phys. 2010, 108. [Google Scholar] [CrossRef]
- Fischer, L.M.; Wilding, N.; Gel, M.; Evoy, S. Low-stress silicon carbonitride for the machining of high-frequency nanomechanical resonators. J. Vac. Sci. Technol. B 2007, 25, 33–37. [Google Scholar] [CrossRef]
- Belov, M.; Quitoriano, N.J.; Sharma, S.; Hiebert, W.K.; Kamins, T.I.; Evoy, S. Mechanical resonance of clamped silicon nanowires measured by optical interferometry. J. Appl. Phys. 2008. [Google Scholar] [CrossRef]
- Quitoriano, N.J.; Belov, M.; Evoy, S.; Kamins, T.I. Single-crystal, Si nanotubes, and their mechanical resonant properties. Nano Lett. 2009, 9, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Solak, A.O.; Eichorst, L.R.; Clark, W.J.; McCreery, R.L. Modified carbon surfaces as “organic electrodes” that exhibit conductance switching. Anal. Chem. 2003, 75, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Weaver, W.; Timoshenko, S.; Young, D.H. Vibration Problems in Engineering, 5th ed.; Wiley: New York, NY, USA, 1990. [Google Scholar]
- Marc, R.E.; Murry, R.F.; Basinger, S.F. Pattern-recognition of amino-acid signatures in retinal neurons. J. Neurosci. 1995, 15, 5106–5129. [Google Scholar] [PubMed]
- Chen, G.Y.; Thundat, T.; Wachter, E.A.; Warmack, R.J. Adsorption-induced surface stress and its effects on resonance frequency of microcantilevers. J. Appl. Phys. 1995, 77, 3618–3622. [Google Scholar] [CrossRef]
- Karabalin, R.B.; Villanueva, L.G.; Matheny, M.H.; Sader, J.E.; Roukes, M.L. Stress-induced variations in the stiffness of micro- and nanocantilever beams. Phys. Rev. Lett. 2012, 108. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, W.; Du, R.; Cao, Y.; Mohammad, M.A.; Dew, S.K.; McDermott, M.T.; Evoy, S. Diazonium Chemistry for the Bio-Functionalization of Glassy Nanostring Resonator Arrays. Sensors 2015, 15, 18724-18741. https://doi.org/10.3390/s150818724
Zheng W, Du R, Cao Y, Mohammad MA, Dew SK, McDermott MT, Evoy S. Diazonium Chemistry for the Bio-Functionalization of Glassy Nanostring Resonator Arrays. Sensors. 2015; 15(8):18724-18741. https://doi.org/10.3390/s150818724
Chicago/Turabian StyleZheng, Wei, Rongbing Du, Yong Cao, Mohammad A. Mohammad, Steven K. Dew, Mark T. McDermott, and Stephane Evoy. 2015. "Diazonium Chemistry for the Bio-Functionalization of Glassy Nanostring Resonator Arrays" Sensors 15, no. 8: 18724-18741. https://doi.org/10.3390/s150818724