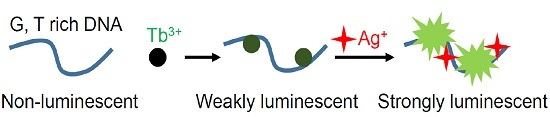
Label-Free Ag+ Detection by Enhancing DNA Sensitized Tb3+ Luminescence
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Enhanced Emission with the Ag10c DNAzyme
3.2. Effect of Ag+ with DNA Homopolymers
3.3. Mechanism Studies
3.4. Sensor Performance
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Drake, P.L.; Hazelwood, K.J. Exposure-related health effects of silver and silver compounds: A review. Ann. Occup. Hyg. 2005, 49, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Ratte, H.T. Bioaccumulation and toxicity of silver compounds: A review. Environ. Toxicol. Chem. 1999, 18, 89–108. [Google Scholar] [CrossRef]
- Coskun, A.; Akkaya, E.U. Ion sensing coupled to resonance energy transfer: A highly selective and sensitive ratiometric fluorescent chemosensor for Ag(I) by a modular approach. J. Am. Chem. Soc. 2005, 127, 10464–10465. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Santra, M.; Won, N.; Kim, S.; Kim, J.K.; Bin Kim, S.; Ahn, K.H. Selective fluorogenic and chromogenic probe for detection of silver ions and silver nanoparticles in aqueous media. J. Am. Chem. Soc. 2009, 131, 2040–2041. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-B.; Kong, R.-M.; Lu, Y. Metal ion sensors based on dnazymes and related DNA molecules. Annu. Rev. Anal. Chem. 2011, 4, 105–128. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, Z.; Lu, Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, K.; Li, Y.F. Biologically inspired synthetic enzymes made from DNA. Chem. Biol. 2009, 16, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Song, S.P.; Qin, Y.; He, Y.; Huang, Q.; Fan, C.H.; Chen, H.Y. Functional nanoprobes for ultrasensitive detection of biomolecules. Chem. Soc. Rev. 2010, 39, 4234–4243. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, R.H.; Yang, L.; Tan, W.H. Nucleic acid conjugated nanomaterials for enhanced molecular recognition. ACS Nano 2009, 3, 2451–2460. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.; Willner, I. Nanobiotechnology: Integrated nanoparticle-biomolecule hybrid systems: Synthesis, properties, and applications. Angew. Chem. Int. Ed. 2004, 43, 6042–6108. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Park, S.H.; Finkelstein, G.; Reif, J.H.; LaBean, T.H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 2003, 301, 1882–1884. [Google Scholar] [CrossRef] [PubMed]
- Keren, K.; Krueger, M.; Gilad, R.; Ben-Yoseph, G.; Sivan, U.; Braun, E. Sequence-specific molecular lithography on single DNA molecules. Science 2002, 297, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Boulais, E.; Hakobyan, Y.; Wang, W.L.; Guan, A.; Bathe, M.; Yin, P. Casting inorganic structures with DNA molds. Science 2014, 346. [Google Scholar] [CrossRef] [PubMed]
- Petty, J.T.; Zheng, J.; Hud, N.V.; Dickson, R.M. DNA-templated Ag nanocluster formation. J. Am. Chem. Soc. 2004, 126, 5207–5212. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, E.G.; O’Neill, P.; Guerrero, A.J.; Bouwmeester, D.; Fygenson, D.K. Sequence-dependent fluorescence of DNA-hosted silver nanoclusters. Adv. Mater. 2008, 20, 279–283. [Google Scholar] [CrossRef]
- Lan, G.Y.; Chen, W.Y.; Chang, H.T. Control of synthesis and optical properties of DNA templated silver nanoclusters by varying DNA length and sequence. RSC Adv. 2011, 1, 802–807. [Google Scholar] [CrossRef]
- Guo, W.; Yuan, J.; Dong, Q.; Wang, E. Highly sequence-dependent formation of fluorescent silver nanoclusters in hybridized DNA duplexes for single nucleotide mutation identification. J. Am. Chem. Soc. 2009, 132, 932–934. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Nicovich, P.R.; Dickson, R.M. Highly fluorescent noble-metal quantum dots. Annu. Rev. Phys. Chem. 2007, 58, 409–431. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Cao, S.; Togashi, H.; Tashiro, M.; Fujimoto, T.; Machinami, T.; Oda, S.; Miyake, Y.; Okamoto, I.; Tanaka, Y. Specific interactions between silver(I) ions and cytosine-cytosine pairs in DNA duplexes. Chem. Commun. 2008, 39, 4825–4827. [Google Scholar] [CrossRef] [PubMed]
- Urata, H.; Yamaguchi, E.; Nakamura, Y.; Wada, S.-I. Pyrimidine-pyrimidine base pairs stabilized by silver(i) ions. Chem. Commun. 2011, 47, 941–943. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Torigoe, H.; Tanaka, Y.; Okamoto, I. Binding of metal ions by pyrimidine base pairs in DNA duplexes. Chem. Soc. Rev. 2011, 40, 5855–5866. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Le, H.-N.; Jiang, X.-Q.; Yin, B.-C.; Ye, B.-C. Time-Resolved Probes Based on Guanine/Thymine-Rich DNA-Sensitized Luminescence of Terbium(III). Anal. Chem. 2013, 85, 11665–11674. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.Q.; Xing, F.F.; He, S.J.; Song, S.P.; Wang, L.H.; Long, Y.T.; Li, D.; Fan, C.H. A graphene-based fluorescent nanoprobe for silver(I) ions detection by using graphene oxide and a silver-specific oligonucleotide. Chem. Commun. 2010, 46, 2596–2598. [Google Scholar] [CrossRef] [PubMed]
- Saran, R.; Liu, J. A Silver DNAzyme. Anal. Chem. 2016, 88, 4014–4020. [Google Scholar] [CrossRef] [PubMed]
- Feig, A.L.; Scott, W.G.; Uhlenbeck, O.C. Inhibition of the hammerhead ribozyme cleavage reaction by site-specific binding of Tb(III). Science 1998, 279, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-K.; Li, J.; Nagraj, N.; Lu, Y. Probing Metal Binding in the 8–17 DNAzyme by TbIII Luminescence Spectroscopy. Chem. Eur. J. 2008, 14, 8696–8703. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ding, J.; Liu, J. A Highly Selective Na+ Aptamer Dissected by Sensitized Tb3+ Luminescence. ChemBioChem 2016, 17, 1563–1570. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.T.D.; Huang, P.-J.J.; Pautler, R.; Liu, J. The group trend of lanthanides binding to DNA and DNAzymes with a complex but symmetric pattern. Chem. Commun. 2014, 50, 11859–11862. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhang, Y.; Huang, P.-J.J.; Ding, J.; Liu, J. A DNAzyme requiring two different metal ions at two distinct sites. Nucleic Acids Res. 2016, 44, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Purohit, C.S.; Verma, S. A Luminescent Silver-Adenine Metallamacrocyclic Quartet. J. Am. Chem. Soc. 2005, 128, 400–401. [Google Scholar] [CrossRef] [PubMed]
- Nabika, H.; Deki, S. Enhancing and Quenching Functions of Silver Nanoparticles on the Luminescent Properties of Europium Complex in the Solution Phase. J. Phys. Chem. B 2003, 107, 9161–9164. [Google Scholar] [CrossRef]
- Tan, H.; Chen, Y. Ag+-enhanced fluorescence of lanthanide/nucleotide coordination polymers and Ag+ sensing. Chem. Commun. 2011, 47, 12373–12375. [Google Scholar] [CrossRef] [PubMed]
- Loo, K.; Degtyareva, N.; Park, J.; Sengupta, B.; Reddish, M.; Rogers, C.C.; Bryant, A.; Petty, J.T. Ag+-Mediated Assembly of 5′-Guanosine Monophosphate. J. Phys. Chem. B 2010, 114, 4320–4326. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Qu, Z.-B.; Ma, H.-Y.; Zhou, T.; Shi, G. DNA-based sensitization of Tb3+ luminescence regulated by Ag+ and cysteine: Use as a logic gate and a H2O2 sensor. Chem. Commun. 2014, 50, 4677–4679. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Chen, X.-Y.; Cheng, P.; Liao, D.-Z.; Yan, S.-P.; Jiang, Z.-H. Coordination Polymers Containing 1D Channels as Selective Luminescent Probes. J. Am. Chem. Soc. 2004, 126, 15394–15395. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Jiao, T.; Li, Y.; Liu, Q.; Tan, M.; Wang, H.; Wang, L. Lanthanide Coordination Polymers and Their Ag+-Modulated Fluorescence. J. Am. Chem. Soc. 2004, 126, 2280–2281. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Tang, G.; Ma, C.; Li, Q. Luminescence detection of cysteine based on Ag+-mediated conformational change of terbium ion-promoted G-quadruplex. Anal. Chim. Acta 2016, 908, 161–167. [Google Scholar] [CrossRef] [PubMed]




© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleinke, K.; Saran, R.; Liu, J. Label-Free Ag+ Detection by Enhancing DNA Sensitized Tb3+ Luminescence. Sensors 2016, 16, 1370. https://doi.org/10.3390/s16091370
Kleinke K, Saran R, Liu J. Label-Free Ag+ Detection by Enhancing DNA Sensitized Tb3+ Luminescence. Sensors. 2016; 16(9):1370. https://doi.org/10.3390/s16091370
Chicago/Turabian StyleKleinke, Kimberly, Runjhun Saran, and Juewen Liu. 2016. "Label-Free Ag+ Detection by Enhancing DNA Sensitized Tb3+ Luminescence" Sensors 16, no. 9: 1370. https://doi.org/10.3390/s16091370