Cadmium-Free Quantum Dots as Fluorescent Labels for Exosomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of InP/ZnS QDs
2.2. Ligand Exchange with MSA and Antibody Conjugation
2.3. Cell Culture
2.4. Fluorescence Microscopy
2.5. Transmission Electron Microscopy
2.6. Nanoparticle Tracking Analysis
2.7. Dynamic Light Scattering
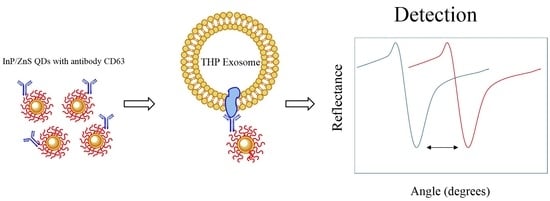
2.8. Surface plasmon resonance (SPR)
2.9. Photoluminescence Spectroscopy
3. Results
3.1. Synthesis and Characterization of the InP/ZnS QDs
3.2. Testing of the InP/ZnS–Anti-CD63 Conjugate
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Medintz, I.L.; Uyeda, H.T.; Goldman, E.R.; Mattoussi, H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 2005, 4, 435–446. [Google Scholar] [CrossRef]
- Boriachek, K.; Islam, M.N.; Gopalan, V.; Lam, A.K.; Nguyen, N.-T.; Shiddiky, M.J.A. Quantum dot-based sensitive detection of disease specific exosome in serum. Analyst 2017, 142, 2211–2219. [Google Scholar] [CrossRef] [PubMed]
- Han, H.-S.; Niemeyer, E.; Huang, Y.; Kamoun, W.S.; Martin, J.D.; Bhaumik, J.; Chen, Y.; Roberge, S.; Cui, J.; Martin, M.R.; et al. Quantum dot/antibody conjugates for in vivo cytometric imaging in mice. Proc. Natl. Acad. Sci. USA 2015, 112, 1350–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zrazhevskiy, P.; Gao, X. Quantum dot imaging platform for single-cell molecular profiling. Nat. Commun. 2013, 4, 1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunetti, V.; Chibli, H.; Fiammengo, R.; Galeone, A.; Malvindi, M.A.; Vecchio, G.; Cingolani, R.; Nadeau, J.L.; Pompa, P.P. InP/ZnS as a safer alternative to CdSe/ZnS core/shell quantum dots: In vitro and in vivo toxicity assessment. Nanoscale 2013, 5, 307–317. [Google Scholar] [CrossRef]
- Boriachek, K.; Islam, M.N.; Möller, A.; Salomon, C.; Nguyen, N.T.; Hossain, M.S.A.; Yamauchi, Y.; Shiddiky, M.J.A. Biological Functions and Current Advances in Isolation and Detection Strategies for Exosome Nanovesicles. Small 2018, 14, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Lobb, R.J.; van Amerongen, R.; Wiegmans, A.; Ham, S.; Larsen, J.E.; Möller, A. Exosomes derived from mesenchymal non-small cell lung cancer cells promote chemoresistance. Int. J. Cancer 2017, 141, 614–620. [Google Scholar] [CrossRef] [Green Version]
- Andreu, Z. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rupert, D.L.M.; Lässer, C.; Eldh, M.; Block, S.; Zhdanov, V.P.; Lotvall, J.O.; Bally, M.; Höök, F. Determination of Exosome Concentration in Solution Using Surface Plasmon Resonance Spectroscopy. Anal. Chem. 2014, 86, 5929–5936. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; EL Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef] [Green Version]
- Van Der Pol, E.; Hoekstra, A.G.; Sturk, A.; Otto, C.; Van Leeuwen, T.G.; Nieuwland, R. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J. Thromb. Haemost. 2010, 8, 2596–2607. [Google Scholar] [CrossRef] [Green Version]
- Van der Pol, E.; Coumans, F.A.W.; Grootemaat, A.E.; Gardiner, C.; Sargent, I.L.; Harrison, P.; Sturk, A.; van Leeuwen, T.G.; Nieuwland, R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J. Thromb. Haemost. 2014, 12, 1182–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.P.; Hole, P.; Carr, B.; Redman, C.W.G.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine 2011, 7, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Tessier, M.D.; Dupont, D.; De Nolf, K.; De Roo, J.; Hens, Z. Economic and Size-tunable Synthesis of InP/ZnE (E = S,Se) Colloidal Quantum Dots. Chem. Mater. 2015, 27, 4893–4898. [Google Scholar] [CrossRef]
- Yong, K.T.; Ding, H.; Roy, I.; Law, W.C.; Bergey, E.J.; Maitra, A.; Prasad, P.N. Imaging pancreatic cancer using bioconjugated inp quantum dots. ACS Nano 2009, 3, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Nann, T. Phase-transfer of CdSe@ZnS quantum dots using amphiphilic hyperbranched polyethylenimine. Chem. Commun. 2005, 1735–1736. [Google Scholar] [CrossRef] [PubMed]
- Trilling, A.K.; Beekwilder, J.; Zuilhof, H. Antibody orientation on biosensor surfaces: A minireview. Analyst 2013, 138, 1619–1627. [Google Scholar] [CrossRef]
- Korpany, K.V.; Mottillo, C.; Bachelder, J.; Cross, S.N.; Dong, P.; Trudel, S.; Friščić, T.; Blum, A.S. One-step ligand exchange and switching from hydrophobic to water-stable hydrophilic superparamagnetic iron oxide nanoparticles by mechanochemical milling. Chem. Commun. 2016, 52, 3054–3057. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Purich, D.L.; Wu, C.; Wu, Y.; Chen, T.; Cui, C.; Zhang, L.; Cansiz, S.; Hou, W.; Wang, Y.; et al. Ionic Functionalization of Hydrophobic Colloidal Nanoparticles To Form Ionic Nanoparticles with Enzymelike Properties. J. Am. Chem. Soc. 2015, 137, 14952–14958. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, R.; Liu, J.; Zhang, B.; Wang, Y.; Liu, X.; Law, W.-C.; Liu, L.; Ye, L.; Yong, K.-T. Cytotoxicity assessment of functionalized CdSe, CdTe and InP quantum dots in two human cancer cell models. Mater. Sci. Eng. C 2015, 57, 222–231. [Google Scholar] [CrossRef]
- Bharali, D.J.; Lucey, D.W.; Jayakumar, H.; Pudavar, H.E.; Prasad, P.N. Folate-Receptor-Mediated Delivery of InP Quantum Dots for Bioimaging Using Confocal and Two-Photon Microscopy. J. Am. Chem. Soc. 2005, 127, 11364–11371. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.; Momen-Heravi, F.; Kodys, K.; Szabo, G. MicroRNA Cargo of Extracellular Vesicles from Alcohol-Exposed Monocytes Signals Naïve Monocytes to Differentiate into M2 Macrophages. J. Biol. Chem. 2015. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zheng, T.; Zhang, B. Exosomes in Parkinson’s Disease. Neurosci. Bull. 2017, 33, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Luo, J.; Jiang, H.; Duan, D.D. Tumor exosomes: A double-edged sword in cancer therapy. Acta Pharmacol. Sin. 2018, 39, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Osada-Oka, M.; Shiota, M.; Izumi, Y.; Nishiyama, M.; Tanaka, M.; Yamaguchi, T.; Sakurai, E.; Miura, K.; Iwao, H. Macrophage-derived exosomes induce inflammatory factors in endothelial cells under hypertensive conditions. Hypertens. Res. 2017, 40, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Sina, A.A.I.; Vaidyanathan, R.; Dey, S.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. Real time and label free profiling of clinically relevant exosomes. Sci. Rep. 2016, 6, 30460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wang, X.; Wang, J.; Fu, W.; Yao, C. A SPR biosensor based on signal amplification using antibody-QD conjugates for quantitative determination of multiple tumor markers. Sci. Rep. 2016, 6, 33140. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.-M.; Quan, L.; Liu, J. Tracking Exosomes in Vitro and in Vivo To Elucidate Their Physiological Functions: Implications for Diagnostic and Therapeutic Nanocarriers. ACS Appl. Nano Mater. 2018, 1, 2438–2448. [Google Scholar] [CrossRef]






| QD Conjugates | Zeta Potential (mV) | Hydrodynamic Diameter (nm) or Size | PL QY (%) | Emission Maximum (nm) | PL FWHM (Full-Width Half-Maximum) | Reference |
|---|---|---|---|---|---|---|
| InP/ZnS–oleylamine | - | 4.96 (DLS) | 37.40% | 600 | 53.00 | This work |
| InP/ZnS–MSA | −37.8 | 4.27 (DLS) | 17.82% | 590 | 55.38 | |
| InP/ZnS–MSA–Anti-CD63 | −7.53 | 86.30 (DLS) | 13.83% | 580 | 70.00 | |
| InP/ZnS–MPA (mercaptopropionic acid) | −26.0 ± 9 | 11.3 ± 0.6 (DLS) | [5] | |||
| InP/ZnS in toluene | 5 ± 0.5 (TEM) | 634 | [20] | |||
| InP/ZnS (with PEGylated phospholipids) | −8.63 | 58.50 (DLS) | ||||
| InP/ZnS in hexane | 4.9–12.6 (DLS) | 15% | 555 | 62.00 | [21] | |
| InP/ZnS–folic acid | 17–24 (DLS) |
| Sample | THP-1 Derived Exosomes | InP/ZnS–Anti-CD63 + THP-1 Derived Exosomes |
|---|---|---|
| Hydrodynamic diameter (Nanosight) | 96.6 ± 7.9 nm | 129.5 ± 20.2 nm |
| Hydrodynamic diameter (DLS) | 36.85 nm | 52.24 nm |
| Size (TEM) | 50–100 nm | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobhal, G.; Ayupova, D.; Laufersky, G.; Ayed, Z.; Nann, T.; Goreham, R.V. Cadmium-Free Quantum Dots as Fluorescent Labels for Exosomes. Sensors 2018, 18, 3308. https://doi.org/10.3390/s18103308
Dobhal G, Ayupova D, Laufersky G, Ayed Z, Nann T, Goreham RV. Cadmium-Free Quantum Dots as Fluorescent Labels for Exosomes. Sensors. 2018; 18(10):3308. https://doi.org/10.3390/s18103308
Chicago/Turabian StyleDobhal, Garima, Deanna Ayupova, Geoffry Laufersky, Zeineb Ayed, Thomas Nann, and Renee V. Goreham. 2018. "Cadmium-Free Quantum Dots as Fluorescent Labels for Exosomes" Sensors 18, no. 10: 3308. https://doi.org/10.3390/s18103308