Development of a Novel Q-body Using an In Vivo Site-Specific Unnatural Amino Acid Incorporation System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Homology Modeling
2.3. Construction of Mutant Antibody and Nef Genes
2.4. Antibody Expression and Purification
2.5. Antigen Expression and Purification
2.6. Fluorescence Measurements
2.7. Western Blotting
2.8. ELISA
3. Results
3.1. Homology Modeling of anti-Nef Antibody
3.2. Construction of the Nef Q-body
3.3. Antigen-dependent Changes in Fluorescence Intensity of the Labeled Antibodies
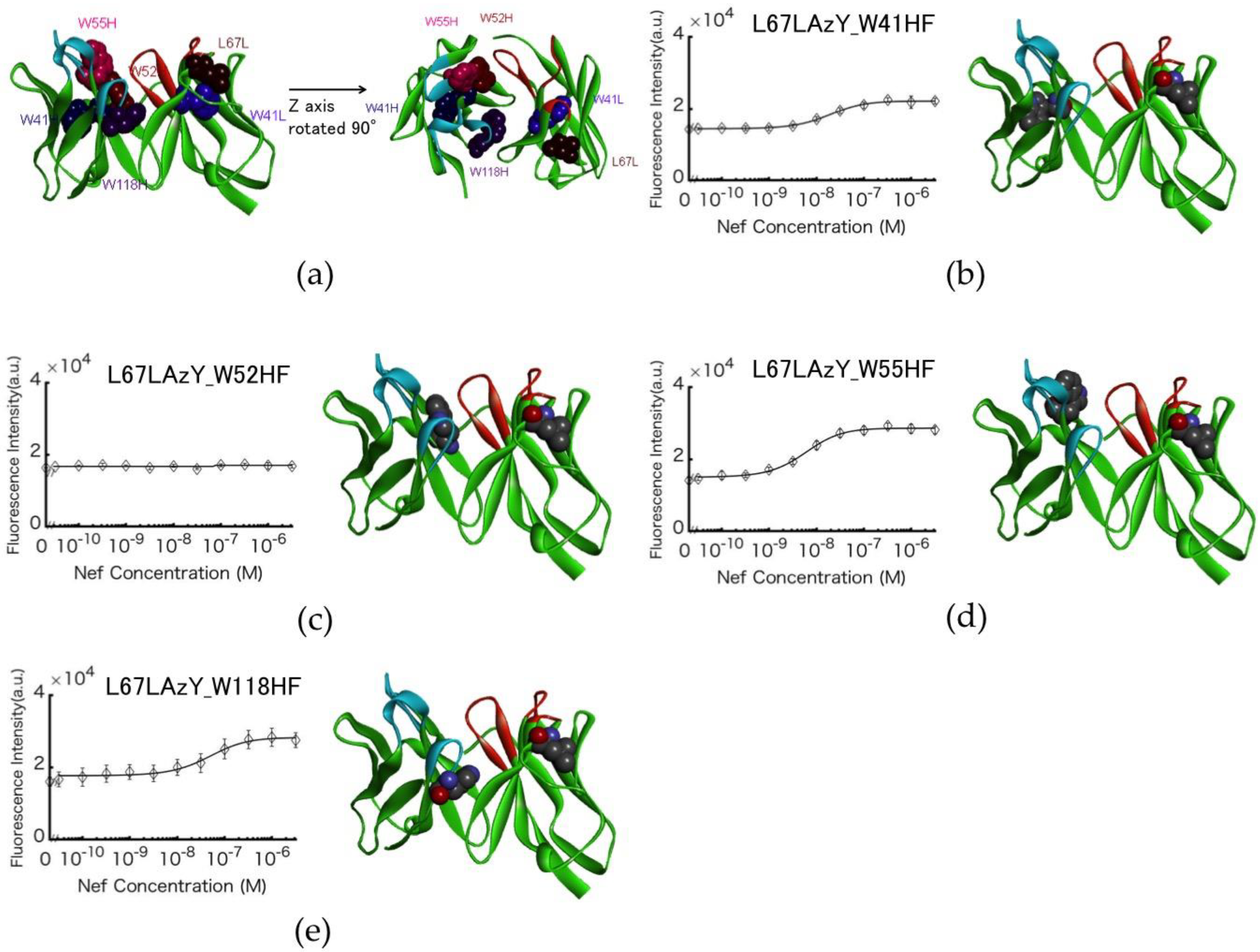
3.4. Involvement of Internal Trp Residues in the Quenching of Cy3 at Position L67L
3.5. Antigen-dependent Changes in Fluorescence Intensity of Labeled Antibodies
3.6. Analysis of the anti-Nef Antibody Epitope
3.7. Decrease in Response Resulting from Mutation of the Antigen (Nef)
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abe, R.; Ohashi, H.; Iijima, I.; Ihara, M.; Takagi, H.; Hohsaka, T.; Ueda, H. “Quenchbodies”: Quench-based antibody probes that show antigen-dependent fluorescence. J. Am. Chem. Soc. 2011, 133, 17386–17394. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.; Dong, J. From fluorescence polarization to Quenchbody: Recent progress in fluorescent reagentless biosensors based on based on antibody and other binding proteins. Biochim. Biophys. Acta 2014, 1844, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Forsstrom, B.; Axnas, B.B.; Rockberg, J.; Danielsson, H.; Bohlin, A.; Uhlen, M. Dissecting antibodies with regards to linear and conformational epitopes. PLoS ONE 2015, 10, e0121673. [Google Scholar] [CrossRef] [PubMed]
- Marme, N.; Knemeyer, J.P.; Sauer, M.; Wolfrum, J. Inter- and intramolecular fluorescence quenching of organic dyes by tryptophan. Bioconjug. Chem. 2003, 14, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- Vaiana, A.C.; Neuweiler, H.; Schulz, A.; Wolfrum, J.; Sauer, M.; Smith, J.C. Fluorescence quenching of dyes by tryptophan: Interactions at atomic detail from combination of experiment and computer simulation. J. Am. Chem. Soc. 2003, 125, 14564–14572. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Ueda, H. Strategy for making a superior Quenchbody to proteins: effect of the fluorophore position. Sensors 2014, 14, 13285–13297. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Kawamura, T.; Dong, J.; Ueda, H. Q-Bodies from Recombinant Single-Chain Fv Fragment with Better Yield and Expanded Palette of Fluorophores. ACS Sens. 2016, 1, 88–94. [Google Scholar] [CrossRef]
- Chalker, J.M.; Bernardes, G.J.; Lin, Y.A.; Davis, B.G. Chemical modification of proteins at cysteine: opportunities in chemistry and biology. Chem. Asian. J. 2009, 4, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Brinkley, M. A brief survey of methods for preparing protein conjugates with dyes, haptens, and cross-linking reagents. Bioconjug. Chem. 1992, 3, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Kojima, T.; Dong, J.; Ohashi, H.; Ueda, H. One-pot construction of Quenchbodies using antibody-binding proteins. Anal. Methods 2016, 43, 7774–7779. [Google Scholar] [CrossRef]
- Abe, R.; Jeong, H.J.; Arakawa, D.; Dong, J.; Ohashi, H.; Kaigome, R.; Saiki, F.; Yamane, K.; Takagi, H.; Ueda, H. Ultra Q-bodies: quench-based antibody probes that utilize dye-dye interactions with enhanced antigen-dependent fluorescence. Sci. Rep. 2014, 4, 4640. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Matsui, M.; Yokogawa, T.; Nakamura, M.; Hosoya, T.; Hiramatsu, T.; Suzuki, M.; Hayashi, N.; Nishikawa, K. Site-selective post-translational modification of proteins using an unnatural amino acid, 3-azidotyrosine. J. Biochem. 2007, 141, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Ikeda-Boku, A.; Ohno, S.; Hibino, Y.; Yokogawa, T.; Hayashi, N.; Nishikawa, K. A simple system for expression of proteins containing 3-azidotyrosine at a pre-determined site in Escherichia coli. J. Biochem. 2013, 153, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Kang, J.A.; Shim, H.E.; Nam, Y.R.; Yoon, S.; Kim, H.R.; Lee, D.E.; Park, S.H. Efficient method for iodine radioisotope labeling of cyclooctyne-containing molecules using strain-promoted copper-free click reaction. Bioorg. Med. Chem. 2015, 23, 3303–3308. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Okuyama, Y.; Hayashi, N. Antibody inhibiting function of HIV Nef through disturbing localization to lipid membrane. (In preparation)
- Fujii, Y.; Nishino, Y.; Nakaya, T.; Tokunaga, K.; Ikuta, K. Expression of human immunodeficiency virus type 1 Nef antigen on the surface of acutely and persistently infected human T cells. Vaccine 1993, 11, 1240–1246. [Google Scholar] [CrossRef]
- Elemento, O.; Lefranc, M.P. IMGT/PhyloGene: An on-line tool for comparative analysis of immunoglobulin and T cell receptor genes. Dev. Comp. Immunol 2003, 27, 763–779. [Google Scholar] [CrossRef]
- RCSB PDB. Available online: http://www.rcsb.org/ (accessed on 4 May 2016).
- Wedemayer, G.J.; Patten, P.A.; Wang, L.H.; Schultz, P.G.; Stevens, R.C. Structural insights into the evolution of an antibody combining site. Science 1997, 276, 1665–1669. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.N.; Hunt, H.D.; Horton, R.M.; Pullen, J.K.; Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989, 77, 51–59. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.30. [Google Scholar]
- Marti-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sanchez, R.; Melo, F.; Sali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.C.; Davis, I.W.; Arendall III, W.B.; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- abYsis. Available online: http://www.bioinf.org.uk/abysis3.1/index.html (accessed on 23 June 2018).
- Chen, H.; Ahsan, S.S.; Santiago-Berrios, B.M.; Abruña, D.H.; Webb, W.W. Mechanisms of Quenching of Alexa Fluorophores by Natural Amino Acids. J. Am. Chem. Soc. 2010, 132, 7244–7245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geyer, M.; Munte, C.E.; Schorr, J.; Kellner, R.; Kalbitzer, H.R. Structure of the anchor-domain of myristoylated and non-myristoylated HIV-1 Nef protein. J. Mol. Biol. 1999, 289, 123–138. [Google Scholar] [CrossRef] [PubMed]






| Primer Name | Nucleotide Sequence (5′-3′) |
|---|---|
| 303_T66LAzY_F | ctacgccgtttcctagttagattctggtgt |
| 303_T66LAzY_R | acaccagaatctaactaggaaacggcgtag |
| 303_L67LAzY_F | cgccgtttccacttaggattctggtgtccc |
| 303_L67LAzY_R | gggacaccagaatcctaagtggaaacggcg |
| 303_S69LAzY_F | ttccactttagattagggtgtcccaaaaag |
| 303_S69LAzY_R | ctttttgggacaccctaatctaaagtggaa |
| 303_D101LAzY_F | tgaagattttgcatagtattactgtctcca |
| 303_D101LAzY_R | tggagacagtaatactatgcaaaatcttca |
| 303_S18LAzY_F | tccatcctccttataggcctctctgggaga |
| 303_S18LAzY_R | tctcccagagaggcctataaggaggatgga |
| 303_W41HF_F | cgactatatgcacttcgtgaagcagaggcc |
| 303_W41HF_R | ggcctctgcttcacgaagtgcatatagtcg |
| 303_W52HF_F | acagggcctggagttcattggatggattga |
| 303_W52HF_R | tcaatccatccaatgaactccaggccctgt |
| 303_W55HF_F | ggagtggattggattcattgatcctgagaa |
| 303_W55HF_R | ttctcaggatcaatgaatccaatccactcc |
| 303_W118H4F_F | tacaagggatgtcttcggcgcagggaccac |
| 303_W118HF_R | gtggtccctgcgccgaagacatcccttgta |
| 303_W41LF_F | tggttacttaagcttccttcagcagaaacc |
| 303_W41LF_R | ggtttctgctgaaggaagcttaagtaacca |
| Nef_del2to11_F | agtagtccatgggatggcctgctgtaagg |
| Nef_R | aagtgtagcggtcacgctgcgcgtaaccac |
| Nef_W13A_F | tagtgtgattggagcacctgctgtaaggga |
| Nef_W13A_R | tcccttacagcaggtgctccaatcacacta |
| Gene | Template | Primers |
|---|---|---|
| T66LAzY | wt anti-Nef antibody | 303_T66LAzY_F, 303_T66LAzY_R |
| L67LAzY | wt anti-Nef antibody | 303_L67LAzY_F, 303_L67LAzY_R |
| S69LAzY | wt anti-Nef antibody | 303_S69LAzY_F, 303_S69LAzY_R |
| D101LAzY | wt anti-Nef antibody | 303_D101LAzY_F, 303_D101LAzY_R |
| S18LAzY | wt anti-Nef antibody | 303_S18LAzY_F, 303_S18LAzY_R |
| L67LAzY_W41HF | L67LAzY | 303_W41HF_F, 303_W41HF_R |
| L67LAzY_W52HF | L67LAzY | 303_W52HF_F, 303_W52HF_R |
| L67LAzY_W55HF | L67LAzY | 303_W55HF_F, 303_W55HF_R |
| L67LAzY_W118HF | L67LAzY | 303_W118HF_F, 303_W118H4F_R |
| L67LAzY_W41LF | L67LAzY | 303_W41LF_F, 303_W41LF_R |
| NefΔ2–10 | Nef wt | Nef_del2to11_F, Nef_R |
| Nef W13A | Nef wt | Nef_W13A_F, Nef_W13A_R |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurumida, Y.; Hayashi, N. Development of a Novel Q-body Using an In Vivo Site-Specific Unnatural Amino Acid Incorporation System. Sensors 2018, 18, 2519. https://doi.org/10.3390/s18082519
Kurumida Y, Hayashi N. Development of a Novel Q-body Using an In Vivo Site-Specific Unnatural Amino Acid Incorporation System. Sensors. 2018; 18(8):2519. https://doi.org/10.3390/s18082519
Chicago/Turabian StyleKurumida, Yoichi, and Nobuhiro Hayashi. 2018. "Development of a Novel Q-body Using an In Vivo Site-Specific Unnatural Amino Acid Incorporation System" Sensors 18, no. 8: 2519. https://doi.org/10.3390/s18082519




