Three-Dimensional High-Resolution Digital Inline Hologram Reconstruction with a Volumetric Deconvolution Method
Abstract
:1. Introduction
2. Materials and Methods
2.1. Optical Imaging Setup
2.2. Sample Preparation
2.3. Hardware and Software for Computation
2.4. Pixel-Super-Resolution (PSR)
2.5. 3D Phase Recovery and Lens-Free Image Reconstruction
2.6. 3D Deconvolution Method
3. Results and Discussion
3.1. Subpixel Super-Resolution and Auto-Focused Z-Stack Reconstruction
3.2. Phase Recovered Hologram Image for Both a Single-Shot and Z-Stacked Image Set
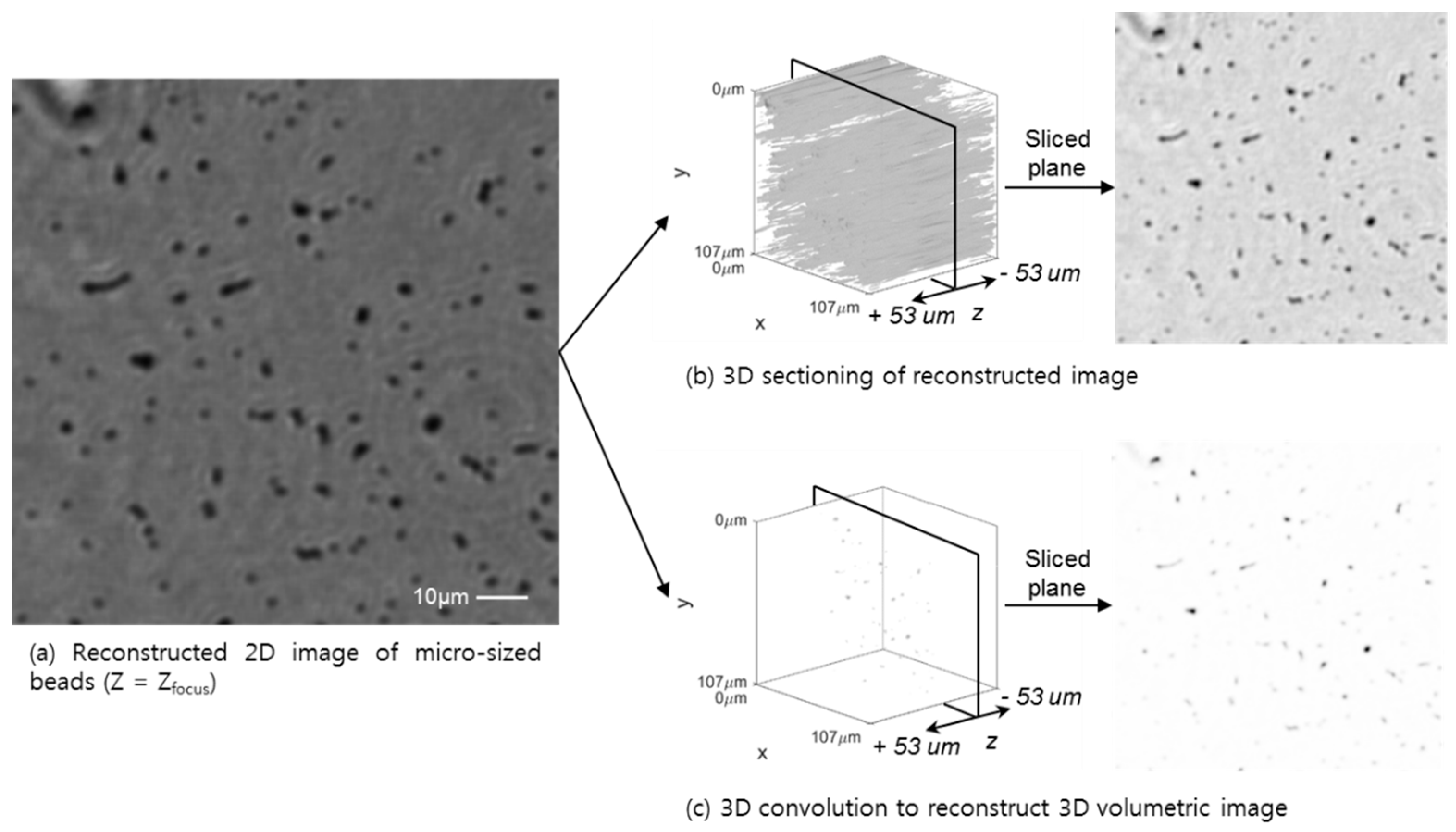
3.3. 3D Sectioning Using the Conventional Auto-Focus Algorithm and 3D Deconvolution Approach
3.4. 3D Deconvolution Approach for a Whole Blood Smear and Biological Tissue Sample
4. Conclusions
Author Contributions
Funding
References
- Gabor, D. A New Microscopic Principle. Nature 1948, 161, 777–778. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Jericho, M.H.; Meinertzhagen, I.A.; Kreuzer, H.J. Digital in-line holography for biological applications. Proc. Natl. Acad. Sci. USA 2001, 98, 11301–11305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.K. Principles and techniques of digital holographic microscopy. Opt. Eng. 2010, 1, 018005. [Google Scholar] [CrossRef]
- Latychevskaia, T.; Fink, H.W. Practical algorithms for simulation and reconstruction of digital in-line holograms. Appl. Opt. 2015, 54, 2424–2434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.K. Digital Holographic Microscopy. In Digital Holographic Microscopy: Principles, Techniques, and Applications; Springer: New York, NY, USA, 2011; pp. 149–190. [Google Scholar]
- Greenbaum, A.; Luo, W.; Su, T.W.; Gorocs, Z.; Xue, L.; Isikman, S.O.; Coskun, A.F.; Mudanyali, O.; Ozcan, A. Imaging without lenses: Achievements and remaining challenges of wide-field on-chip microscopy. Nat. Methods 2012, 9, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Vashist, S.K.; Luppa, P.B.; Yeo, L.Y.; Ozcan, A.; Luong, J.H.T. Emerging Technologies for Next-Generation Point-of-Care Testing. Trends Biotechnol. 2015, 33, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, A.; McLeod, E. Lensless Imaging and Sensing. Annu. Rev. Biomed. Eng. 2016, 18, 77–102. [Google Scholar] [CrossRef] [PubMed]
- Latychevskaia, T.; Fink, H.W. Coherent microscopy at resolution beyond diffraction limit using post-experimental data extrapolation. Appl. Phys. Lett. 2013, 103, 204105. [Google Scholar] [CrossRef] [Green Version]
- Kanka, M.; Riesenberg, R.; Petruck, P.; Graulig, C. High resolution (NA = 0.8) in lensless in-line holographic microscopy with glass sample carriers. Opt. Lett. 2011, 36, 3651–3653. [Google Scholar] [CrossRef] [PubMed]
- Bishara, W.; Su, T.W.; Coskun, A.F.; Ozcan, A. Lensfree on-chip microscopy over a wide field-of-view using pixel super-resolution. Opt. Express 2010, 18, 11181–11191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, C.; Isikman, S.O.; Khademhosseinieh, B.; Ozcan, A. On-chip differential interference contrast microscopy using lensless digital holography. Opt. Express 2010, 18, 4717–4726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coskun, A.F.; Su, T.W.; Ozcan, A. Wide field-of-view lens-free fluorescent imaging on a chip. Lab Chip 2010, 10, 824–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.B.; Shin, Y.; Sung, K.; Yang, S.; Chen, H.; Wang, H.D.; Teng, D.; Rivenson, Y.; Kulkarni, R.P.; Ozcan, A. 3D imaging of optically cleared tissue using a simplified CLARITY method and on-chip microscopy. Sci. Adv. 2017, 3, e1700553. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Hong, J.S.; Liu, C.G.; Kim, M.K. Review of digital holographic microscopy for three-dimensional profiling and tracking. Opt. Eng. 2014, 53, 112306. [Google Scholar] [CrossRef] [Green Version]
- Memmolo, P.; Miccio, L.; Paturzo, M.; Di Caprio, G.; Coppola, G.; Netti, P.A.; Ferraro, P. Recent advances in holographic 3D particle tracking. Adv. Opt. Photonics 2015, 7, 713–755. [Google Scholar] [CrossRef]
- Isikman, S.O.; Bishara, W.; Sikora, U.; Yaglidere, O.; Yeah, J.; Ozcan, A. Field-portable lensfree tomographic microscope. Lab Chip 2011, 11, 2222–2230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Jeon, P.; Kim, D. 3D image distortion problem in digital in-line holographic microscopy and its effective solution. Opt. Express 2017, 25, 21969–21980. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, A.; Zhang, Y.B.; Feizi, A.; Chung, P.L.; Luo, W.; Kandukuri, S.R.; Ozcan, A. Wide-field computational imaging of pathology slides using lens-free on-chip microscopy. Sci. Transl. Med. 2014, 6, 267ra175. [Google Scholar] [CrossRef] [PubMed]
- Latychevskaia, T.; Longchamp, J.N.; Fink, H.W. Novel Fourier-domain constraint for fast phase retrieval in coherent diffraction imaging. Opt. Express 2011, 19, 19330–19339. [Google Scholar] [CrossRef] [PubMed]
- Latychevskaia, T.; Fink, H.W. Reconstruction of purely absorbing, absorbing and phase-shifting, and strong phase-shifting objects from their single-shot in-line holograms. Appl. Opt. 2015, 54, 3925–3932. [Google Scholar] [CrossRef]
- Su, T.W.; Isikman, S.O.; Bishara, W.; Tseng, D.; Erlinger, A.; Ozcan, A. Multi-angle lensless digital holography for depth resolved imaging on a chip. Opt. Express 2010, 18, 9690–9711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Sun, J.; Chen, Q.; Li, J.; Zuo, C. Adaptive pixel-super-resolved lensfree in-line digital holography for wide-field on-chip microscopy. Sci. Rep. 2017, 7, 11777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenbaum, A.; Luo, W.; Khademhosseinieh, B.; Su, T.W.; Coskun, A.F.; Ozcan, A. Increased space-bandwidth product in pixel super-resolved lensfree on-chip microscopy. Sci. Rep. 2013, 3, 1717. [Google Scholar] [CrossRef]
- Garcia-Sucerquia, J. Color lensless digital holographic microscopy with micrometer resolution. Opt. Lett. 2012, 37, 1724–1726. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, A.; Feizi, A.; Akbari, N.; Ozcan, A. Wide-field computational color imaging using pixel super-resolved on-chip microscopy. Opt. Express 2013, 21, 12469–12483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.K.; Naik, D.N.; Pedrini, G.; Takeda, M.; Osten, W. Exploiting scattering media for exploring 3D objects. Light-Sci. Appl. 2017, 6, e16219. [Google Scholar] [CrossRef] [PubMed]
- Antipa, N.; Kuo, G.; Heckel, R.; Mildenhall, B.; Bostan, E.; Ng, R.; Waller, L. DiffuserCam: Lensless single-exposure 3D imaging. Optica 2018, 5, 1–9. [Google Scholar] [CrossRef]
- Mukherjee, S.; Vijayakumar, A.; Kumar, M.; Rosen, J. 3D Imaging through Scatterers with Interferenceless Optical System. Sci. Rep. 2018, 8, 1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Zhang, Y.; Feizi, A.; Gorocs, Z.; Ozcan, A. Pixel super-resolution using wavelength scanning. Light-Sci. Appl. 2016, 5, e16060. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Zhang, Y.B.; Gorocs, Z.; Feizi, A.; Ozcan, A. Propagation phasor approach for holographic image reconstruction. Sci. Rep. 2016, 6, 22738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheong, F.C.; Sun, B.; Dreyfus, R.; Amato-Grill, J.; Xiao, K.; Dixon, L.; Grier, D.G. Flow visualization and flow cytometry with holographic video microscopy. Opt. Express 2009, 17, 13071–13079. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Hong, J.; Liu, C.G.; Cross, M.; Haynie, D.T.; Kim, M.K. Four-dimensional motility tracking of biological cells by digital holographic microscopy. J. Biomed. Opt. 2014, 19, 045001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latychevskaia, T.; Gehri, F.; Fink, H.W. Depth-resolved holographic reconstructions by three-dimensional deconvolution. Opt. Express 2010, 18, 22527–22544. [Google Scholar] [CrossRef] [PubMed]
- Latychevskaia, T.; Fink, H.W. Holographic time-resolved particle tracking by means of three-dimensional volumetric deconvolution. Opt. Express 2014, 22, 20994–21003. [Google Scholar] [CrossRef] [PubMed]
- Bishara, W.; Sikora, U.; Mudanyali, O.; Su, T.W.; Yaglidere, O.; Luckhart, S.; Ozcan, A. Holographic pixel super-resolution in portable lensless on-chip microscopy using a fiber-optic array. Lab Chip 2011, 11, 1276–1279. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, A.; Sikora, U.; Ozcan, A. Field-portable wide-field microscopy of dense samples using multi-height pixel super-resolution based lensfree imaging. Lab Chip 2012, 12, 1242–1245. [Google Scholar] [CrossRef] [PubMed]
- Rivenson, Y.; Gorocs, Z.; Gunaydin, H.; Zhang, Y.B.; Wang, H.D.; Ozcan, A. Deep learning microscopy. Optica 2017, 4, 1437–1443. [Google Scholar] [CrossRef]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eom, J.; Moon, S. Three-Dimensional High-Resolution Digital Inline Hologram Reconstruction with a Volumetric Deconvolution Method. Sensors 2018, 18, 2918. https://doi.org/10.3390/s18092918
Eom J, Moon S. Three-Dimensional High-Resolution Digital Inline Hologram Reconstruction with a Volumetric Deconvolution Method. Sensors. 2018; 18(9):2918. https://doi.org/10.3390/s18092918
Chicago/Turabian StyleEom, Junseong, and Sangjun Moon. 2018. "Three-Dimensional High-Resolution Digital Inline Hologram Reconstruction with a Volumetric Deconvolution Method" Sensors 18, no. 9: 2918. https://doi.org/10.3390/s18092918




