Evaluation of Molecularly Imprinted Polymers for Point-of-Care Testing for Cardiovascular Disease
Abstract
1. Introduction
2. Protein Detection
2.1. Challenges of Protein Detection Using PIPs
2.2. Heterogeneity
2.3. Binding Kinetics
2.4. Solvent Compatability
3. Detection Techniques
3.1. Sandwich Complexes
3.2. Stimuli-Responsive Species
3.3. Signal Quenching
3.4. Classic ‘Label-Free’ Techniques
3.5. Multiplexed Detection
3.6. Feasability for Point-of-Care Testing
4. Biomimetic Sensors Analytical Performance
4.1. Evaluating Comparitive Performance
4.2. Clinical Considerations
5. MIPs for Point-of-Care Testing
Cardiovascular Risk Stratification
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wieser, S.; Riguzzi, M.; Pletscher, M.; Huber, C.A.; Telser, H.; Schwenkglenks, M. How Much Does the Treatment of Each Major Disease Cost? A Decomposition of Swiss National Health Accounts. Eur. J. Health Econ. 2018, 19, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Dieleman, J.L.; Baral, R.; Birger, M.; Bui, A.L.; Bulchis, A.; Chapin, A.; Hamavid, H.; Horst, C.; Johnson, E.K.; Joseph, J.; et al. US Spending on Personal Health Care and Public Health, 1996-2013. JAMA 2016, 316, 2627–2646. [Google Scholar] [CrossRef] [PubMed]
- Timmis, A.; Townsend, N.; Gale, C.; Grobbee, R.; Maniadakis, N.; Flather, M.; Wilkins, E.; Wright, L.; Vos, R.; Bax, J.; et al. European Society of Cardiology: Cardiovascular Disease Statistics 2017. Eur. Heart J. 2018, 39, 508–579. [Google Scholar] [CrossRef] [PubMed]
- Arboleda, V.A.; Garner, O.B. Ensuring the Quality of Point-of-Care Testing in a Large and Decentralized Ambulatory Care Setting. Am. J. Clin. Pathol. 2017, 148, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.L.V. Practical Challenges Related to Point of Care Testing. Pract. Lab. Med. 2016, 4, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Bingisser, R.; Cairns, C.; Christ, M.; Hausfater, P.; Lindahl, B.; Mair, J.; Panteghini, M.; Price, C.; Venge, P. Cardiac Troponin: A Critical Review of the Case for Point-of-Care Testing in the ED. Am. J. Emerg. Med. 2012, 30, 1639–1649. [Google Scholar] [CrossRef] [PubMed]
- Florkowski, C.; Don-Wauchope, A.; Gimenez, N.; Rodriguez-Capote, K.; Wils, J.; Zemlin, A. Point-of-Care Testing (POCT) and Evidence-Based Laboratory Medicine (EBLM)—Does It Leverage Any Advantage in Clinical Decision Making? Crit. Rev. Clin. Lab. Sci. 2017. [Google Scholar] [CrossRef]
- Hsia, R.Y.; Hale, Z.; Tabas, J.A. A National Study of the Prevalence of Life-Threatening Diagnoses in Patients With Chest Pain. JAMA Intern. Med. 2016, 176, 1029–1032. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.J.; Ardise, J.; Gulla, J.; Cangro, J. Point-of-Care Testing Reduces Length of Stay in Emergency Department Chest Pain Patients. Ann. Emerg. Med. 2005, 45, 587–591. [Google Scholar] [CrossRef]
- Renaud, B.; Maison, P.; Ngako, A.; Cunin, P.; Santin, A.; Hervé, J.; Salloum, M.; Calmettes, M.-J.; Boraud, C.; Lemiale, V.; et al. Impact of Point-of-Care Testing in the Emergency Department Evaluation and Treatment of Patients with Suspected Acute Coronary Syndromes. Acad. Emerg. Med. Off. J. Soc. Acad. Emerg. Med. 2008, 15, 216–224. [Google Scholar] [CrossRef]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Chaitman, B.R.; Bax, J.J.; Morrow, D.A.; White, H.D.; The Executive Group on behalf of the Joint European Society of Cardiology (ESC)/American College of Cardiology (ACC)/American Heart Association (AHA)/World Heart Federation (WHF) Task Force for the Universal Definition of Myocardial Infarction. Fourth Universal Definition of Myocardial Infarction. Circulation 2018, 138, e618–e651. [Google Scholar] [CrossRef] [PubMed]
- IFCC C-CB. High-Sensitivity* Cardiac Troponin I and T Assay Analytical Characteristics Designated by Manufacturer; IFCC Committee on Clinical Applications of Cardiac Bio-Markers (C-CB): Milan, Italy, 2018. [Google Scholar]
- IFCC C-CB. Point of Care Cardiac Troponin I and T Assay Analytical Characteristics Designated by Manufacturer; IFCC Committee on Clinical Applications of Cardiac Bio-Markers (C-CB): Milan, Italy, 2018. [Google Scholar]
- Aldous, S.; Mark Richards, A.; George, P.M.; Cullen, L.; Parsonage, W.A.; Flaws, D.; Florkowski, C.M.; Troughton, R.W.; O’Sullivan, J.W.; Reid, C.M.; et al. Comparison of New Point-of-Care Troponin Assay with High Sensitivity Troponin in Diagnosing Myocardial Infarction. Int. J. Cardiol. 2014, 177, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Pickering, J.W.; Young, J.M.; George, P.M.; Watson, A.S.; Aldous, S.J.; Troughton, R.W.; Pemberton, C.J.; Richards, A.M.; Cullen, L.A.; Than, M.P. Validity of a Novel Point-of-Care Troponin Assay for Single-Test Rule-Out of Acute Myocardial Infarction. JAMA Cardiol. 2018, 3, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Body, R.; Almashali, M.; Morris, N.; Moss, P.; Jarman, H.; Appelboam, A.; Parris, R.; Chan, L.; Walker, A.; Harrison, M.; et al. Diagnostic Accuracy of the T-MACS Decision Aid with a Contemporary Point-of-Care Troponin Assay. Heart 2019, 105, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Juliano, M.; Wason, C. Comparison of Point-of-Care Versus Laboratory Troponin Testing in an Emergency Department Setting. Mil. Med. 2017, 182, e1938–e1940. [Google Scholar] [CrossRef] [PubMed]
- Christenson, R.H.; Mullins, K.; Duh, S.-H. Validation of High-Sensitivity Performance for a United States Food and Drug Administration Cleared Cardiac Troponin I Assay. Clin. Biochem. 2018, 56, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Masoum, S. Molecularly Imprinted Polymers for Capturing and Sensing Proteins: Current Progress and Future Implications. TrAC Trends Anal. Chem. 2019, 114, 29–47. [Google Scholar] [CrossRef]
- Verheyen, E.; Schillemans, J.P.; van Wijk, M.; Demeniex, M.-A.; Hennink, W.E.; van Nostrum, C.F. Challenges for the Effective Molecular Imprinting of Proteins. Biomaterials 2011, 32, 3008–3020. [Google Scholar] [CrossRef]
- Nigam, P.K. Biochemical Markers of Myocardial Injury. Indian J. Clin. Biochem. 2007, 22, 10–17. [Google Scholar] [CrossRef]
- McDonnell, B.; Hearty, S.; Leonard, P.; O’Kennedy, R. Cardiac Biomarkers and the Case for Point-of-Care Testing. Clin. Biochem. 2009, 42, 549–561. [Google Scholar] [CrossRef]
- Mayes, A.G.; Whitcombe, M.J. Synthetic Strategies for the Generation of Molecularly Imprinted Organic Polymers. Adv. Drug Deliv. Rev. 2005, 57, 1742–1778. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, M.; Mori, T.; Ariga, K. Molecular Imprinting: Materials Nanoarchitectonics with Molecular Information. Bull. Chem. Soc. Jpn. 2018, 91, 1075–1111. [Google Scholar] [CrossRef]
- Chen, W.; Ma, Y.; Pan, J.; Meng, Z.; Pan, G.; Sellergren, B. Molecularly Imprinted Polymers with Stimuli-Responsive Affinity: Progress and Perspectives. Polymers 2015, 7, 1689–1715. [Google Scholar] [CrossRef]
- Su, X.; Li, X.; Li, J.; Liu, M.; Lei, F.; Tan, X.; Li, P.; Luo, W. Synthesis and Characterization of Core–shell Magnetic Molecularly Imprinted Polymers for Solid-Phase Extraction and Determination of Rhodamine B in Food. Food Chem. 2015, 171, 292–297. [Google Scholar] [CrossRef]
- Turner, A.P.F. Biosensors: Sense and Sensibility. Chem. Soc. Rev. 2013, 42, 3184–3196. [Google Scholar] [CrossRef] [PubMed]
- Ndunda, E.N.; Mizaikoff, B. Molecularly Imprinted Polymers for the Analysis and Removal of Polychlorinated Aromatic Compounds in the Environment: A Review. Analyst 2016, 141, 3141–3156. [Google Scholar] [CrossRef] [PubMed]
- Walshe, M.; Howarth, J.; Kelly, M.T.; O’Kennedy, R.; Smyth, M.R. The Preparation of a Molecular Imprinted Polymer to 7- Hydroxycoumarin and Its Use as a Solid-Phase Extraction Material. J. Pharm. Biomed. Anal. 1997, 16, 319–325. [Google Scholar] [CrossRef]
- Liu, M.; Ding, X.; Yang, Q.; Wang, Y.; Zhao, G.; Yang, N. A PM Leveled Photoelectrochemical Sensor for Microcystin-LR Based on Surface Molecularly Imprinted TiO2@CNTs Nanostructure. J. Hazard. Mater. 2017, 331, 309–320. [Google Scholar] [CrossRef]
- Zhao, X.; Hu, W.; Wang, Y.; Zhu, L.; Yang, L.; Sha, Z.; Zhang, J. Decoration of Graphene with 2-Aminoethanethiol Functionalized Gold Nanoparticles for Molecular Imprinted Sensing of Erythrosine. Carbon 2018, 127, 618–626. [Google Scholar] [CrossRef]
- Han, X.; Li, S.; Peng, Z.; Othman, A.M.; Leblanc, R. Recent Development of Cardiac Troponin I Detection. ACS Sens. 2016, 1, 106–114. [Google Scholar] [CrossRef]
- Bakirhan, N.K.; Ozcelikay, G.; Ozkan, S.A. Recent Progress on the Sensitive Detection of Cardiovascular Disease Markers by Electrochemical-Based Biosensors. J. Pharm. Biomed. Anal. 2018, 159, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Maurine, M.M. The Point-of-Care Diagnostic Landscape for Sexually Transmitted Infections (STIs); World Health Organisation: Geneva, Switzerland, 2018. [Google Scholar]
- Kettler, H.; White, K.; Hawkes, S. TDR | Mapping the Landscape of Diagnostics for Sexually Transmitted Infections; World Health Organisation: Geneva, Switzerland, 2004; p. 44. [Google Scholar]
- St John, A.; Price, C.P. Economic Evidence and Point-of-Care Testing. Clin. Biochem. Rev. 2013, 34, 61–74. [Google Scholar] [PubMed]
- Luppa, P.B.; Junker, R.; Langer, C. Definitions and Areas of Application. In Point-of-Care Testing: Principles and Clinical Applications; Luppa, P.B., Junker, R., Eds.; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2018; pp. 3–7. [Google Scholar]
- Eriksson, S.; Junikka, M.; Pettersson, K. An Interfering Component in Cardiac Troponin I Immunoassays-Its Nature and Inhibiting Effect on the Binding of Antibodies against Different Epitopes. Clin. Biochem. 2004, 37, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Hyytiä, H.; Heikkilä, T.; Hedberg, P.; Puolakanaho, T.; Pettersson, K. Skeletal Troponin I Cross-Reactivity in Different Cardiac Troponin I Assay Versions. Clin. Biochem. 2015, 48, (4–5). [Google Scholar] [CrossRef] [PubMed]
- Savukoski, T.; Engström, E.; Engblom, J.; Ristiniemi, N.; Wittfooth, S.; Lindahl, B.; Eggers, K.M.; Venge, P.; Pettersson, K. Troponin-Specific Autoantibody Interference in Different Cardiac Troponin I Assay Configurations. Clin. Chem. 2012, 58, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Peronnet, E.; Becquart, L.; Poirier, F.; Cubizolles, M.; Choquet-Kastylevsky, G.; Jolivet-Reynaud, C. SELDI-TOF MS Analysis of the Cardiac Troponin I Forms Present in Plasma from Patients with Myocardial Infarction. Proteomics 2006, 6, 6288–6299. [Google Scholar] [CrossRef] [PubMed]
- Labugger, R.; Organ, L.; Collier, C.; Atar, D.; Van Eyk, J.E. Extensive Troponin I and T Modification Detected in Serum From Patients With Acute Myocardial Infarction. Circulation 2000, 102, 1221–1226. [Google Scholar] [CrossRef] [PubMed]
- Apple, F.S.; Ler, R.; Murakami, M.M. Determination of 19 Cardiac Troponin I and T Assay 99th Percentile Values from a Common Presumably Healthy Population. Clin. Chem. 2012, 58, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Mair, J.; Lindahl, B.; Müller, C.; Giannitsis, E.; Huber, K.; Möckel, M.; Plebani, M.; Thygesen, K.; Jaffe, A.S. What to Do When You Question Cardiac Troponin Values. Eur. Heart J. Acute Cardiovasc. Care 2018, 7, 577–586. [Google Scholar] [CrossRef]
- Schmid, J.; Liesinger, L.; Birner-Gruenberger, R.; Stojakovic, T.; Scharnagl, H.; Dieplinger, B.; Asslaber, M.; Radl, R.; Beer, M.; Polacin, M.; et al. Elevated Cardiac Troponin T in Patients With Skeletal Myopathies. J. Am. Coll. Cardiol. 2018, 71, 1540–1549. [Google Scholar] [CrossRef]
- Tsui, A.K.Y.; Lyon, M.E.; van Diepen, S.; Goudreau, B.L.; Thomas, D.; Higgins, T.; Raizman, J.E.; Füzéry, A.K.; Rodriguez-Capote, K.; Estey, M.; et al. Analytical Concordance of Diverse Point-of-Care and Central Laboratory Troponin I Assays. J. Appl. Lab. Med. 2018. [Google Scholar] [CrossRef]
- Bates, K.J.; Hall, E.M.; Fahie-Wilson, M.N.; Kindler, H.; Bailey, C.; Lythall, D.; Lamb, E.J. Circulating Immunoreactive Cardiac Troponin Forms Determined by Gel Filtration Chromatography after Acute Myocardial Infarction. Clin. Chem. 2010, 56, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, R.; Wang, H.; Li, J.; Qu, L.; Harrington, P. High-Selective and Sensitive Voltammetric Sensor for Butylated Hydroxyanisole Based on AuNPs–PVP–graphene Nanocomposites. Talanta 2015, 138, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Liu, Y.; Yan, Z.; Liu, L. Molecularly Imprinted Solid Phase Extraction Coupled to High Performance Liquid Chromatography for Determination of Aflatoxin M1 and B1 in Foods and Feeds. RSC Adv. 2015, 5, 20951–20960. [Google Scholar] [CrossRef]
- Ali, W.H.; Derrien, D.; Alix, F.; Pérollier, C.; Lépine, O.; Bayoudh, S.; Chapuis-Hugon, F.; Pichon, V. Solid-Phase Extraction Using Molecularly Imprinted Polymers for Selective Extraction of a Mycotoxin in Cereals. J. Chromatogr. A 2010, 1217, 6668–6673. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, S.; Li, J. Recent Advances in Molecular Imprinting Technology: Current Status, Challenges and Highlighted Applications. Chem. Soc. Rev. 2011, 40, 2922–2942. [Google Scholar] [CrossRef] [PubMed]
- Ansell, R.J. Characterization of the Binding Properties of Molecularly Imprinted Polymers. Adv. Biochem. Eng. Biotechnol. 2015, 150, 51–93. [Google Scholar] [CrossRef] [PubMed]
- Schauperl, M.; Lewis, D.W. Probing the Structural and Binding Mechanism Heterogeneity of Molecularly Imprinted Polymers. J. Phys. Chem. B 2015, 119, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Ashley, J.; Feng, X.; Halder, A.; Zhou, T.; Sun, Y. Dispersive Solid-Phase Imprinting of Proteins for the Production of Plastic Antibodies. Chem. Commun. 2018, 54, 3355–3358. [Google Scholar] [CrossRef]
- Gao, R.; Hao, Y.; Zhang, L.; Cui, X.; Liu, D.; Zhang, M.; Tang, Y.; Zheng, Y. A Facile Method for Protein Imprinting on Directly Carboxyl-Functionalized Magnetic Nanoparticles Using Non-Covalent Template Immobilization Strategy. Chem. Eng. J. 2016, 284, 139–148. [Google Scholar] [CrossRef]
- Nishino, H.; Huang, C.-S.; Shea, K.J. Selective Protein Capture by Epitope Imprinting. Angew. Chem. Int. Ed. 2006, 45, 2392–2396. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ambrosini, S.; Tamahkar, E.; Rossi, C.; Haupt, K.; Tse Sum Bui, B. Toward a Universal Method for Preparing Molecularly Imprinted Polymer Nanoparticles with Antibody-like Affinity for Proteins. Biomacromolecules 2016, 17, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.T.C.; Dutra, R.A.F.; Noronha, J.P.C.; Cunha, A.L.; Sales, M.G.F. Artificial Antibodies for Troponin T by Its Imprinting on the Surface of Multiwalled Carbon Nanotubes: Its Use as Sensory Surfaces. Biosens. Bioelectron. 2011, 28, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.; Feng, Y.J.; Contois, J.H.; Pervaiz, S. Comparison of Myoglobin, Creatine Kinase-MB, and Cardiac Troponin I for Diagnosis of Acute Myocardial Infarction. Ann. Clin. Lab. Sci. 1996, 26, 291–300. [Google Scholar] [PubMed]
- Wittenberg, B.A.; Wittenberg, J.B.; Caldwell, P.R. Role of Myoglobin in the Oxygen Supply to Red Skeletal Muscle. J. Biol. Chem. 1975, 250, 9038–9043. [Google Scholar]
- Sallach, S.M.; Nowak, R.; Hudson, M.P.; Tokarski, G.; Khoury, N.; Tomlanovich, M.C.; Jacobsen, G.; de Lemos, J.A.; McCord, J. A Change in Serum Myoglobin to Detect Acute Myocardial Infarction in Patients with Normal Troponin I Levels. Am. J. Cardiol. 2004, 94, 864–867. [Google Scholar] [CrossRef]
- Moreira, F.T.C.; Dutra, R.A.F.; Noronha, J.P.C.; Fernandes, J.C.S.; Sales, M.G.F. Novel Biosensing Device for Point-of-Care Applications with Plastic Antibodies Grown on Au-Screen Printed Electrodes. Sens. Actuators B Chem. 2013, 182, 733–740. [Google Scholar] [CrossRef][Green Version]
- Jordanova, N.; Gyöngyösi, M.; Khorsand, A.; Falkensammer, C.; Zorn, G.; Wojta, J.; Anvari, A.; Huber, K. New Cut-off Values of Cardiac Markers for Risk Stratification of Angina Pectoris. Int. J. Cardiol. 2005, 99, 429–435. [Google Scholar] [CrossRef]
- Zayats, M.; Brenner, A.J.; Searson, P.C. Protein Imprinting in Polyacrylamide-Based Gels. Biomaterials 2014, 35, 8659–8668. [Google Scholar] [CrossRef][Green Version]
- Yarman, A.; Jetzschmann, K.J.; Neumann, B.; Zhang, X.; Wollenberger, U.; Cordin, A.; Haupt, K.; Scheller, F.W. Enzymes as Tools in MIP-Sensors. Chemosensors 2017, 5, 11. [Google Scholar] [CrossRef]
- Moreira, F.T.C.; Sharma, S.; Dutra, R.A.F.; Noronha, J.P.C.; Cass, A.E.G.; Sales, M.G.F. Protein-Responsive Polymers for Point-of-Care Detection of Cardiac Biomarker. Sens. Actuators B Chem. 2014, 196, 123–132. [Google Scholar] [CrossRef]
- Erdőssy, J.; Kassa, E.; Farkas, A.; Horváth, V. Enzymatic Digestion as a Tool for Removing Proteinaceous Templates from Molecularly Imprinted Polymers. Anal. Methods 2017, 9, 4496–4503. [Google Scholar] [CrossRef]
- Venton, D.L.; Gudipati, E. Influence of Protein on Polysiloxane Polymer Formation: Evidence for Induction of Complementary Protein-Polymer Interactions. Biochim. Biophys. Acta 1995, 1250, 126–136. [Google Scholar] [CrossRef]
- Ribeiro, J.A.; Pereira, C.M.; Silva, A.F.; Sales, M.G.F. Electrochemical Detection of Cardiac Biomarker Myoglobin Using Polyphenol as Imprinted Polymer Receptor. Anal. Chim. Acta 2017, 981, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhi, K.; Zhang, Y.; Liu, Y.; Zhang, L.; Yasin, A.; Lin, Q. Molecularly Imprinted Polymers for Gossypol via Sol–Gel, Bulk, and Surface Layer Imprinting—A Comparative Study. Polymers 2019, 11, 602. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Tan, T.; Svec, F. Molecular Imprinting of Proteins in Polymers Attached to the Surface of Nanomaterials for Selective Recognition of Biomacromolecules. Biotechnol. Adv. 2013, 31, 1172–1186. [Google Scholar] [CrossRef]
- Sharma, P.S.; Pietrzyk-Le, A.; D’Souza, F.; Kutner, W. Electrochemically Synthesized Polymers in Molecular Imprinting for Chemical Sensing. Anal. Bioanal. Chem. 2012, 402, 3177–3204. [Google Scholar] [CrossRef] [PubMed]
- Blanco-López, M.C.; Gutiérrez-Fernández, S.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Electrochemical Sensing with Electrodes Modified with Molecularly Imprinted Polymer Films. Anal. Bioanal. Chem. 2004, 378, 1922–1928. [Google Scholar] [CrossRef]
- Shumyantseva, V.V.; Bulko, T.V.; Sigolaeva, L.V.; Kuzikov, A.V.; Archakov, A.I. Electrosynthesis and Binding Properties of Molecularly Imprinted Poly-o-Phenylenediamine for Selective Recognition and Direct Electrochemical Detection of Myoglobin. Biosens. Bioelectron. 2016, 86, 330–336. [Google Scholar] [CrossRef]
- Liu, J.; Deng, Q.; Tao, D.; Yang, K.; Zhang, L.; Liang, Z.; Zhang, Y. Preparation of Protein Imprinted Materials by Hierarchical Imprinting Techniques and Application in Selective Depletion of Albumin from Human Serum. Sci. Rep. 2014, 4, 5487. [Google Scholar] [CrossRef]
- Titirici, M.M.; Sellergren, B. Peptide Recognition via Hierarchical Imprinting. Anal. Bioanal. Chem. 2004, 378, 1913–1921. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, M.; Hayatsu, Y.; Nakata, H.; Ishii, Y.; Ito, R.; Saito, K.; Nakazawa, H. Molecularly Imprinted Solid Phase Extraction Using Stable Isotope Labeled Compounds as Template and Liquid Chromatography–mass Spectrometry for Trace Analysis of Bisphenol A in Water Sample. Anal. Chim. Acta 2005, 539, 83–89. [Google Scholar] [CrossRef]
- Chou, P.-C.; Rick, J.; Chou, T.-C. C-Reactive Protein Thin-Film Molecularly Imprinted Polymers Formed Using a Micro-Contact Approach. Anal. Chim. Acta 2005, 542, 20–25. [Google Scholar] [CrossRef]
- Zethelius, B.; Berglund, L.; Sundström, J.; Ingelsson, E.; Basu, S.; Larsson, A.; Venge, P.; Arnlöv, J. Use of Multiple Biomarkers to Improve the Prediction of Death from Cardiovascular Causes. N. Engl. J. Med. 2008, 358, 2107–2116. [Google Scholar] [CrossRef] [PubMed]
- Panduranga, P.; Riyami, A.A.; Sulaiman, K.J.; Mukhaini, M. C-Reactive Protein in Unstable Angina: Clinical and Angiographic Correlation. Heart Asia 2010, 2, 140–144. [Google Scholar] [CrossRef]
- Fonseca, F.A.H.; de Oliveira Izar, M.C. High-Sensitivity C-Reactive Protein and Cardiovascular Disease Across Countries and Ethnicities. Clinics 2016, 71, 235–242. [Google Scholar] [CrossRef]
- Cortez, A.F.; Muxfeldt, E.S.; Cardoso, C.R.L.; Salles, G.F. Prognostic Value of C-Reactive Protein in Resistant Hypertension. Am. J. Hypertens. 2016, 29, 992–1000. [Google Scholar] [CrossRef]
- Nakaya, T.; Li, Y. Recent Progress of Phospholipid Polymers. Des. Monomers Polym. 2003, 6, 309–351. [Google Scholar] [CrossRef]
- Lawton, J.M.; Habib, M.; Ma, B.; Brooks, R.A.; Best, S.M.; Lewis, A.L.; Rushton, N.; Bonfield, W. The Effect of Cationically-Modified Phosphorylcholine Polymers on Human Osteoblasts in Vitro and Their Effect on Bone Formation in Vivo. J. Mater. Sci. Mater. Med. 2017, 28, 144. [Google Scholar] [CrossRef]
- Liu, X.; Xiao, T.; Wu, F.; Shen, M.-Y.; Zhang, M.; Yu, H.; Mao, L. Ultrathin Cell-Membrane-Mimic Phosphorylcholine Polymer Film Coating Enables Large Improvements for In Vivo Electrochemical Detection. Angew. Chem. Int. Ed. 2017, 56, 11802–11806. [Google Scholar] [CrossRef]
- Jin, Y.J.; Kang, S.; Park, P.; Choi, D.; Kim, D.W.; Jung, D.; Koh, J.; Jeon, J.; Lee, M.; Ham, J.; et al. Anti-Inflammatory and Antibacterial Effects of Covalently Attached Biomembrane-Mimic Polymer Grafts on Gore-Tex Implants. ACS Appl. Mater. Interfaces 2017, 9, 19161–19175. [Google Scholar] [CrossRef] [PubMed]
- Hammouda, B.; Horkay, F.; Becker, M.L. Clustering and Solvation in Poly(Acrylic Acid) Polyelectrolyte Solutions. Macromolecules 2005, 38, 2019–2021. [Google Scholar] [CrossRef]
- Duan, F.; Chen, C.; Zhao, X.; Yang, Y.; Liu, X.; Qin, Y. Water-Compatible Surface Molecularly Imprinted Polymers with Synergy of Bi-Functional Monomers for Enhanced Selective Adsorption of Bisphenol A from Aqueous Solution. Environ. Sci. Nano 2016, 3, 213–222. [Google Scholar] [CrossRef]
- Figueiredo, K.C.S.; Ferraz, H.C.; Borges, C.P.; Alves, T.L.M. Structural Stability of Myoglobin in Organic Media. Protein J. 2009, 28, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Treviño, S.; Prabhakaran, E.; Scholtz, J.M. Protein Structure, Stability and Solubility in Water and Other Solvents. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 1225–1235. [Google Scholar] [CrossRef]
- Sullivan, M.V.; Dennison, S.R.; Archontis, G.; Reddy, S.M.; Hayes, J.M. Toward Rational Design of Selective Molecularly Imprinted Polymers (MIPs) for Proteins: Computational and Experimental Studies of Acrylamide Based Polymers for Myoglobin. J. Phys. Chem. B 2019, 123, 5432–5443. [Google Scholar] [CrossRef] [PubMed]
- Boroznjak, R.; Reut, J.; Tretjakov, A.; Lomaka, A.; Öpik, A.; Syritski, V. A Computational Approach to Study Functional Monomer-Protein Molecular Interactions to Optimize Protein Molecular Imprinting. J. Mol. Recognit. 2017, 30, e2635. [Google Scholar] [CrossRef]
- Kryscio, D.R.; Shi, Y.; Ren, P.; Peppas, N.A. Molecular Docking Simulations for Macromolecularly Imprinted Polymers. Ind. Eng. Chem. Res. 2011, 50, 13877–13884. [Google Scholar] [CrossRef] [PubMed]
- Whitcombe, M.J.; Chianella, I.; Larcombe, L.; Piletsky, S.A.; Noble, J.; Porter, R.; Horgan, A. The Rational Development of Molecularly Imprinted Polymer-Based Sensors for Protein Detection. Chem. Soc. Rev. 2011, 40, 1547–1571. [Google Scholar] [CrossRef]
- Turner, N.W.; Jeans, C.W.; Brain, K.R.; Allender, C.J.; Hlady, V.; Britt, D.W. From 3D to 2D: A Review of the Molecular Imprinting of Proteins. Biotechnol. Prog. 2006, 22, 1474–1489. [Google Scholar] [CrossRef]
- Kryscio, D.R.; Peppas, N.A. Critical Review and Perspective of Macromolecularly Imprinted Polymers. Acta Biomater. 2012, 8, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Bossi, A.M.; Sharma, P.S.; Montana, L.; Zoccatelli, G.; Laub, O.; Levi, R. Fingerprint-Imprinted Polymer: Rational Selection of Peptide Epitope Templates for the Determination of Proteins by Molecularly Imprinted Polymers. Anal. Chem. 2012, 84, 4036–4041. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.; Ludman, A.J.; Dworzynski, K.; Al-Mohammad, A.; Cowie, M.R.; McMurray, J.J.V.; Mant, J.; NICE Guideline Development Group for Acute Heart Failure. The Diagnostic Accuracy of the Natriuretic Peptides in Heart Failure: Systematic Review and Diagnostic Meta-Analysis in the Acute Care Setting. BMJ 2015, 350, h910. [Google Scholar] [CrossRef] [PubMed]
- Dietl, A.; Stark, K.; Zimmermann, M.E.; Meisinger, C.; Schunkert, H.; Birner, C.; Maier, L.S.; Peters, A.; Heid, I.M.; Luchner, A. NT-ProBNP Predicts Cardiovascular Death in the General Population Independent of Left Ventricular Mass and Function: Insights from a Large Population-Based Study with Long-Term Follow-Up. PLoS ONE 2016, 11, e0164060. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Li, D.; Ma, J.; Shan, L.; Wei, M. NT-ProBNP Test with Improved Accuracy for the Diagnosis of Chronic Heart Failure. Medicine 2017, 96. [Google Scholar] [CrossRef] [PubMed]
- Ponikowski, P.; Voors, A.A.; Anker, S.D.; Bueno, H.; Cleland, J.G.F.; Coats, A.J.S.; Falk, V.; González-Juanatey, J.R.; Harjola, V.-P.; Jankowska, E.A.; et al. 2016 ESC Guidelines for the Diagnosis and Treatment of Acute and Chronic Heart Failure: The Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure of the European Society of Cardiology (ESC)Developed with the Special Contribution of the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 2016, 37, 2129–2200. [Google Scholar] [CrossRef] [PubMed]
- Regan, B.; O’Kennedy, R.; Collins, D. Point-of-Care Compatibility of Ultra-Sensitive Detection Techniques for the Cardiac Biomarker Troponin I—Challenges and Potential Value. Biosensors 2018, 8, 114. [Google Scholar] [CrossRef]
- Amundson, B.E.; Apple, F.S. Cardiac Troponin Assays: A Review of Quantitative Point-of-Care Devices and Their Efficacy in the Diagnosis of Myocardial Infarction. Clin. Chem. Lab. Med. 2015, 53, 665–676. [Google Scholar] [CrossRef]
- Hayden, O. One Binder to Bind Them All. Sensors 2016, 16. [Google Scholar] [CrossRef]
- Hongzhi, L.; Shoufang, X. Functional Monomer-Template-QDs Sandwich Structure for Mesoporous Structured Bovine Hemoglobin Imprinted Ratiometric Fluorescence Sensor. Talanta 2017, 165, 482–488. [Google Scholar] [CrossRef]
- You, M.; Yang, S.; Tang, W.; Zhang, F.; He, P.-G. Ultrasensitive Electrochemical Detection of Glycoprotein Based on Boronate Affinity Sandwich Assay and Signal Amplification with Functionalized SiO2@Au Nanocomposites. ACS Appl. Mater. Interfaces 2017, 9, 13855–13864. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gan, N.; Hu, F.; Li, T.; Zhou, H.; Li, X.; Zheng, L. A Single Antibody Sandwich Electrochemiluminescence Immunosensor Based on Protein Magnetic Molecularly Imprinted Polymers Mimicking Capture Probes. Sens. Actuators B Chem. 2013, 186, 300–307. [Google Scholar] [CrossRef]
- Lee, M.-H.; Thomas, J.L.; Chen, Y.-C.; Chin, W.-T.; Lin, H.-Y. The Complete Replacement of Antibodies by Protein-Imprinted Poly(Ethylene-Co-Vinyl Alcohol) in Sandwich Fluoroimmunoassays. Microchim. Acta 2013, 180, 1393–1399. [Google Scholar] [CrossRef]
- Kim, E.; Kim, H.-C.; Lee, S.G.; Lee, S.J.; Go, T.-J.; Baek, C.S.; Jeong, S.W. C-Reactive Protein-Directed Immobilization of Phosphocholine Ligands on a Solid Surface. Chem. Commun. Camb. Engl. 2011, 47, 11900–11902. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Goldys, E.M.; Liu, G. Molecularly Imprinted Polymer-Based Reusable Biosensing Device on Stainless Steel for Spatially Localized Detection of Cytokine IL-1β. Sens. Actuators B Chem. 2019, 292, 277–283. [Google Scholar] [CrossRef]
- Pascual-Figal, D.A.; Bayes-Genis, A.; Asensio-Lopez, M.C.; Hernández-Vicente, A.; Garrido-Bravo, I.; Pastor-Perez, F.; Díez, J.; Ibáñez, B.; Lax, A. The Interleukin-1 Axis and Risk of Death in Patients With Acutely Decompensated Heart Failure. J. Am. Coll. Cardiol. 2019, 73, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Correia, L.C.L.; Andrade, B.B.; Borges, V.M.; Clarêncio, J.; Bittencourt, A.P.; Freitas, R.; Souza, A.C.; Almeida, M.C.; Leal, J.; Esteves, J.P.; et al. Prognostic Value of Cytokines and Chemokines in Addition to the GRACE Score in Non-ST-Elevation Acute Coronary Syndromes. Clin. Chim. Acta 2010, 411, 540–545. [Google Scholar] [CrossRef]
- Bujak, M.; Frangogiannis, N.G. The Role of Interleukin-1 in the Pathogenesis of Heart Disease. Arch. Immunol. Ther. Exp. 2009, 57, 165–176. [Google Scholar] [CrossRef]
- Monnerat, G.; Alarcón, M.L.; Vasconcellos, L.R.; Hochman-Mendez, C.; Brasil, G.; Bassani, R.A.; Casis, O.; Malan, D.; Travassos, L.H.; Sepúlveda, M.; et al. Macrophage-Dependent IL-1β Production Induces Cardiac Arrhythmias in Diabetic Mice. Nat. Commun. 2016, 7, 13344. [Google Scholar] [CrossRef]
- Peiró, C.; Lorenzo, Ó.; Carraro, R.; Sánchez-Ferrer, C.F. IL-1β Inhibition in Cardiovascular Complications Associated to Diabetes Mellitus. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef]
- Hasdai, D.; Scheinowitz, M.; Leibovitz, E.; Sclarovsky, S.; Eldar, M.; Barak, V. Increased Serum Concentrations of Interleukin-1 Beta in Patients with Coronary Artery Disease. Heart 1996, 76, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Guo, Q.; Cao, C.; Yang, H.; Li, B. Thermo-Responsive Molecularly Imprinted Nanogels for Specific Recognition and Controlled Release of Proteins. Soft Matter 2013, 9, 3840–3850. [Google Scholar] [CrossRef]
- Miyata, T.; Jige, M.; Nakaminami, T.; Uragami, T. Tumor Marker-Responsive Behavior of Gels Prepared by Biomolecular Imprinting. Proc. Natl. Acad. Sci. USA 2006, 103, 1190–1193. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.; Gariano, N.A.; Spivak, D.A. Macromolecular Amplification of Binding Response in Superaptamer Hydrogels. J. Am. Chem. Soc. 2013, 135, 6977–6984. [Google Scholar] [CrossRef] [PubMed]
- Filipcsei, G.; Fehér, J.; Zrínyi, M. Electric Field Sensitive Neutral Polymer Gels. J. Mol. Struct. 2000, 554, 109–117. [Google Scholar] [CrossRef]
- Wan, W.; Wagner, S.; Rurack, K. Fluorescent Monomers: “Bricks” That Make a Molecularly Imprinted Polymer “Bright”. Anal. Bioanal. Chem. 2016, 408, 1753–1771. [Google Scholar] [CrossRef]
- Sunayama, H.; Ooya, T.; Takeuchi, T. Fluorescent Protein Recognition Polymer Thin Films Capable of Selective Signal Transduction of Target Binding Events Prepared by Molecular Imprinting with a Post-Imprinting Treatment. Biosens. Bioelectron. 2010, 26, 458–462. [Google Scholar] [CrossRef]
- Turan, E.; Ozçetin, G.; Caykara, T. Dependence of Protein Recognition of Temperature-Sensitive Imprinted Hydrogels on Preparation Temperature. Macromol. Biosci. 2009, 9, 421–428. [Google Scholar] [CrossRef]
- Ahmed, E.M. Hydrogel: Preparation, Characterization, and Applications: A Review. J. Adv. Res. 2015, 6, 105–121. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Y.; Mateescu, A.; Sergelen, K.; Kibrom, A.; Jonas, U.; Wei, T.; Dostalek, J. Biosensor Based on Hydrogel Optical Waveguide Spectroscopy for the Detection of 17β-Estradiol. Talanta 2013, 104, 149–154. [Google Scholar] [CrossRef]
- Koetting, M.C.; Peters, J.T.; Steichen, S.D.; Peppas, N.A. Stimulus-Responsive Hydrogels: Theory, Modern Advances, and Applications. Mater. Sci. Eng. R Rep. 2015, 93, 1–49. [Google Scholar] [CrossRef] [PubMed]
- Dolak, İ.; Keçili, R.; Onat, R.; Ziyadanoğulları, B.; Ersöz, A.; Say, R. Molecularly Imprinted Affinity Cryogels for the Selective Recognition of Myoglobin in Blood Serum. J. Mol. Struct. 2018, 1174, 171–176. [Google Scholar] [CrossRef]
- Ertürk, G.; Bereli, N.; Ramteke, P.W.; Denizli, A. Molecularly Imprinted Supermacroporous Cryogels for Myoglobin Recognition. Appl. Biochem. Biotechnol. 2014, 173, 1250–1262. [Google Scholar] [CrossRef] [PubMed]
- Muslumova, S.; Yetiskin, B.; Okay, O. Highly Stretchable and Rapid Self-Recoverable Cryogels Based on Butyl Rubber as Reusable Sorbent. Gels 2019, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Bandow, J.E. Comparison of Protein Enrichment Strategies for Proteome Analysis of Plasma. PROTEOMICS 2010, 10, 1416–1425. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.E.; Lyass, A.; Courchesne, P.; Chen, G.; Liu, C.; Yin, X.; Hwang, S.-J.; Massaro, J.M.; Larson, M.G.; Levy, D. Protein Biomarkers of Cardiovascular Disease and Mortality in the Community. J. Am. Heart Assoc. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-D.; Huang, Q.-W.; Ma, C.; Liu, X.-Y.; Zhang, H.-X. Magnetic Fluorescent Molecularly Imprinted Nanoparticles for Detection and Separation of Transferrin in Human Serum. Talanta 2018, 188, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Stack, A.G.; Mutwali, A.I.; Nguyen, H.T.; Cronin, C.J.; Casserly, L.F.; Ferguson, J. Transferrin Saturation Ratio and Risk of Total and Cardiovascular Mortality in the General Population. QJM Int. J. Med. 2014, 107, 623–633. [Google Scholar] [CrossRef]
- Ruhe, J.; Waldeyer, C.; Ojeda, F.; Altay, A.; Schnabel, R.B.; Schäfer, S.; Lackner, K.J.; Blankenberg, S.; Zeller, T.; Karakas, M. Intrinsic Iron Release Is Associated with Lower Mortality in Patients with Stable Coronary Artery Disease—First Report on the Prospective Relevance of Intrinsic Iron Release. Biomolecules 2018, 8. [Google Scholar] [CrossRef]
- Stack, A.G.; Mohamed, W.; Elsayed, M. Transferrin Saturation Ratio: A Method to Estimate Risk of Cardiovascular Mortality in the General Population? Biomark. Med. 2014, 8, 913–915. [Google Scholar] [CrossRef]
- Piloto, A.M.; Ribeiro, D.S.M.; Rodrigues, S.S.M.; Santos, C.; Santos, J.L.M.; Sales, M.G.F. Plastic Antibodies Tailored on Quantum Dots for an Optical Detection of Myoglobin down to the Femtomolar Range. Sci. Rep. 2018, 8, 4944. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Li, C.; Zhang, C. Development of Quantum Dot-Based Biosensors: Principles and Applications. J. Mater. Chem. B 2018, 6, 6173–6190. [Google Scholar] [CrossRef]
- Yang, Z.; Fan, J.Z.; Proppe, A.H.; de Arquer, F.P.G.; Rossouw, D.; Voznyy, O.; Lan, X.; Liu, M.; Walters, G.; Quintero-Bermudez, R.; et al. Mixed-Quantum-Dot Solar Cells. Nat. Commun. 2017, 8, 1325. [Google Scholar] [CrossRef] [PubMed]
- Melanson, S.F.; Lewandrowski, E.L.; Januzzi, J.L.; Lewandrowski, K.B. Reevaluation of Myoglobin for Acute Chest Pain Evaluation: Would False-Positive Results on “First-Draw” Specimens Lead to Increased Hospital Admissions? Am. J. Clin. Pathol. 2004, 121, 804–808. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, M.; Ye, X.; Wu, K.; Wu, T.; Li, C. Voltammetric Myoglobin Sensor Based on a Glassy Carbon Electrode Modified with a Composite Film Consisting of Carbon Nanotubes and a Molecularly Imprinted Polymerized Ionic Liquid. Microchim. Acta 2017, 184, 195–202. [Google Scholar] [CrossRef]
- Livi, S.; Duchet-Rumeau, J.; Gérard, J.-F.; Pham, T.N. Polymers and Ionic Liquids: A Successful Wedding. Macromol. Chem. Phys. 2015, 216, 359–368. [Google Scholar] [CrossRef]
- Sardar, J.; Krasnou, I.; Baddam, V.; Gudkova, V.; Krumme, A.; Savest, N.; Tarasova, E.; Viirsalu, M.; Mäeorg, U. Synthesis of Polymerizable Ionic Liquid Monomer and Its Characterization. IOP Conf. Ser. Mater. Sci. Eng. Online 2016, 111. [Google Scholar] [CrossRef]
- Palladino, P.; Minunni, M.; Scarano, S. Cardiac Troponin T Capture and Detection in Real-Time via Epitope-Imprinted Polymer and Optical Biosensing. Biosens. Bioelectron. 2018, 106, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Erickson, D. Surface Plasmon Resonance Sensors. In Encyclopedia of Microfluidics and Nanofluidics; Li, D., Ed.; Springer US: Boston, MA, USA, 2008; pp. 1939–1945. [Google Scholar]
- McRae, A.; Graham, M.; Abedin, T.; Ji, Y.; Yang, H.; Wang, D.; Southern, D.; Andruchow, J.; Lang, E.; Innes, G.; et al. Sex-Specific, High-Sensitivity Cardiac Troponin T Cut-off Concentrations for Ruling out Acute Myocardial Infarction with a Single Measurement. CJEM 2019, 21, 26–33. [Google Scholar] [CrossRef]
- Vashist, S.K.; Vashist, P. Recent Advances in Quartz Crystal Microbalance-Based Sensors. J. Sens. 2011, 2011, 13. [Google Scholar] [CrossRef]
- Deakin, M.R.; Buttry, D.A. Electrochemical Applications of the Quartz Crystal Microbalance. Anal. Chem. 1989, 61, 1147A–1154A. [Google Scholar] [CrossRef]
- Lieberzeit, P.A.; Chunta, S.; Navakul, K.; Sangma, C.; Jungmann, C. Molecularly Imprinted Polymers for Diagnostics: Sensing High Density Lipoprotein and Dengue Virus. Procedia Eng. 2016, 168, 101–104. [Google Scholar] [CrossRef]
- Keene, D.; Price, C.; Shun-Shin, M.J.; Francis, D.P. Effect on Cardiovascular Risk of High Density Lipoprotein Targeted Drug Treatments Niacin, Fibrates, and CETP Inhibitors: Meta-Analysis of Randomised Controlled Trials Including 117 411 Patients. BMJ 2014, 349, g4379. [Google Scholar] [CrossRef] [PubMed]
- Santos-Gallego, C.G.; Badimón, J.J.; Santos-Gallego, C.G.; Badimón, J.J. High-Density Lipoprotein and Cardiovascular Risk Reduction, Promises and Realities. Rev. Esp. Cardiol. 2012, 65, 305–308. [Google Scholar] [CrossRef]
- Silbernagel, G.; Schöttker, B.; Appelbaum, S.; Scharnagl, H.; Kleber, M.E.; Grammer, T.B.; Ritsch, A.; Mons, U.; Holleczek, B.; Goliasch, G.; et al. High-Density Lipoprotein Cholesterol, Coronary Artery Disease, and Cardiovascular Mortality. Eur. Heart J. 2013, 34, 3563–3571. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.T.C.; Dutra, R.A.F.; Noronha, J.P.C.; Sales, M.G.F. Electrochemical Biosensor Based on Biomimetic Material for Myoglobin Detection. Electrochimica Acta 2013, 107, 481–487. [Google Scholar] [CrossRef]
- Apple, F.S.; Smith, S.W.; Pearce, L.A.; Murakami, M.M. Assessment of the Multiple-Biomarker Approach for Diagnosis of Myocardial Infarction in Patients Presenting with Symptoms Suggestive of Acute Coronary Syndrome. Clin. Chem. 2009, 55, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Ocak, T.; Erdem, A.; Duran, A.; Tekelioğlu, Ü.Y.; Öztürk, S.; Ayhan, S.S.; Özlü, M.F.; Tosun, M.; Koçoğlu, H.; Yazıcı, M. The Diagnostic Significance of NT-ProBNP and Troponin I in Emergency Department Patients Presenting with Palpitations. Clinics 2013, 68, 543–547. [Google Scholar] [CrossRef]
- Hoffmann, U.; Espeter, F.; Weiß, C.; Ahmad-Nejad, P.; Lang, S.; Brueckmann, M.; Akin, I.; Neumaier, M.; Borggrefe, M.; Behnes, M. Ischemic Biomarker Heart-Type Fatty Acid Binding Protein (HFABP) in Acute Heart Failure - Diagnostic and Prognostic Insights Compared to NT-ProBNP and Troponin I. BMC Cardiovasc. Disord. 2015, 15. [Google Scholar] [CrossRef]
- Panagiotopoulou, M.; Salinas, Y.; Beyazit, S.; Kunath, S.; Duma, L.; Prost, E.; Mayes, A.G.; Resmini, M.; Tse Sum Bui, B.; Haupt, K. Molecularly Imprinted Polymer Coated Quantum Dots for Multiplexed Cell Targeting and Imaging. Angew. Chem. 2016, 128, 8384–8388. [Google Scholar] [CrossRef]
- Ensafi, A.A.; Nasr-Esfahani, P.; Rezaei, B. Simultaneous Detection of Folic Acid and Methotrexate by an Optical Sensor Based on Molecularly Imprinted Polymers on Dual-Color CdTe Quantum Dots. Anal. Chim. Acta 2017, 996, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Xu, G.; Wu, Y.; Wang, X.; Yang, J.; Liu, L.; Zhou, P.; Hu, Q. Molecularly Imprinted Polymers on Dual-Color Quantum Dots for Simultaneous Detection of Norepinephrine and Epinephrine. Sens. Actuators B Chem. 2016, 229, 38–46. [Google Scholar] [CrossRef]
- Feng, F.; Zheng, J.; Qin, P.; Han, T.; Zhao, D. A Novel Quartz Crystal Microbalance Sensor Array Based on Molecular Imprinted Polymers for Simultaneous Detection of Clenbuterol and Its Metabolites. Talanta 2017, 167, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Si, B.; Song, E. Molecularly Imprinted Polymers for the Selective Detection of Multi-Analyte Neurotransmitters. Microelectron. Eng. 2018, 187–188, 58–65. [Google Scholar] [CrossRef]
- Stejskal, J.; Bober, P.; Trchová, M.; Nuzhnyy, D.; Bovtun, V.; Savinov, M.; Petzelt, J.; Prokeš, J. Interfaced Conducting Polymers. Synth. Met. 2017, 224, 109–115. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Y.; Zuo, J.; Zhang, J.; Zhu, L.; Zhang, J. Rapid and Sensitive Determination of Tartrazine Using a Molecularly Imprinted Copolymer Modified Carbon Electrode (MIP-PmDB/PoPD-GCE). J. Electroanal. Chem. 2017, 785, 90–95. [Google Scholar] [CrossRef]
- Lv, Y.; Qin, Y.; Svec, F.; Tan, T. Molecularly Imprinted Plasmonic Nanosensor for Selective SERS Detection of Protein Biomarkers. Biosens. Bioelectron. 2016, 80, 433–441. [Google Scholar] [CrossRef]
- Yee, M.F.; Emmel, G.N.; Yang, E.J.; Lee, E.; Paek, J.H.; Wu, B.M.; Kamei, D.T. Ionic Liquid Aqueous Two-Phase Systems for the Enhanced Paper-Based Detection of Transferrin and Escherichia Coli. Front. Chem. 2018, 6. [Google Scholar] [CrossRef]
- Mayang, Y.; He, X.; Chen, L.; Zhang, Y. Detection of Transferrin by Using a Surface Plasmon Resonance Sensor Functionalized with a Boronic Acid Monolayer. Microchim. Acta 2017, 184, 2749–2757. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, Y.; Xu, B.; Zhang, H.; Gao, Y.; Zhang, H.; Song, D. A Novel Surface Plasmon Resonance Biosensor Based on Graphene Oxide Decorated with Gold Nanorod-Antibody Conjugates for Determination of Transferrin. Biosens. Bioelectron. 2013, 45, 230–236. [Google Scholar] [CrossRef]
- Lam, T. A New Era in Affordable Raman Spectroscopy. Raman Technol. Today’s Spectrosc. 2004, 1, 30–37. [Google Scholar]
- Pence, I.; Mahadevan-Jansen, A. Clinical Instrumentation and Applications of Raman Spectroscopy. Chem. Soc. Rev. 2016, 45, 1958–1979. [Google Scholar] [CrossRef] [PubMed]
- Blacksberg, J.; Alerstam, E.; Maruyama, Y.; Cochrane, C.J.; Rossman, G.R. Miniaturized Time-Resolved Raman Spectrometer for Planetary Science Based on a Fast Single Photon Avalanche Diode Detector Array. Appl. Opt. 2016, 55, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Blacksberg, J.; Maruyama, Y.; Charbon, E.; Rossman, G.R. Fast Single-Photon Avalanche Diode Arrays for Laser Raman Spectroscopy. Opt. Lett. 2011, 36, 3672–3674. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, S.; Akkus, O. A Customized Raman System for Point-of-Care Detection of Arthropathic Crystals in the Synovial Fluid. Analyst 2014, 139, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Li, Y.; Li, Y.; Li, S.; Li, S.; Zhong, J.; Zhong, J. Portable Surface Plasmon Resonance Biosensor and Its Applications. In International Conference on Photonics and Imaging in Biology and Medicine, paper W3A.23; Optical Society of America: Washington, DC, USA, 2017; p. W3A.23. [Google Scholar] [CrossRef]
- Guner, H.; Ozgur, E.; Kokturk, G.; Celik, M.; Esen, E.; Topal, A.E.; Ayas, S.; Uludag, Y.; Elbuken, C.; Dana, A. A Smartphone Based Surface Plasmon Resonance Imaging (SPRi) Platform for on-Site Biodetection. Sens. Actuators B Chem. 2017, 239, 571–577. [Google Scholar] [CrossRef]
- Šípová, H.; Piliarik, M.; Vala, M.; Chadt, K.; Adam, P.; Bocková, M.; Hegnerová, K.; Homola, J. Portable Surface Plasmon Resonance Biosensor for Detection of Nucleic Acids. Procedia Eng. 2011, 25, 148–151. [Google Scholar] [CrossRef][Green Version]
- O’Reilly, E.J.; Conroy, P.J.; Hearty, S.; Keyes, T.E.; O’Kennedy, R.; Forster, R.J.; Dennany, L. Electrochemiluminescence Platform for the Detection of C-Reactive Proteins: Application of Recombinant Antibody Technology to Cardiac Biomarker Detection. RSC Adv. 2015, 5, 67874–67877. [Google Scholar] [CrossRef]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. ScFv Antibody: Principles and Clinical Application. Clin. Dev. Immunol. 2012, 2012, 980250. [Google Scholar] [CrossRef]
- Hearty, S.; O’Kennedy, R. Exploiting Recombinant Antibodies in Point-of-Care (POC) Diagnostics: The Combinatorial Advantage. Bioeng. Bugs 2011, 2, 182–186. [Google Scholar] [CrossRef]
- Bowen, J.L.; Manesiotis, P.; Allender, C.J. Twenty Years since ‘Antibody Mimics’ by Molecular Imprinting Were First Proposed: A Critical Perspective. Mol. Impr. 2013, 1, 35–40. [Google Scholar] [CrossRef][Green Version]
- Canfarotta, F.; Cecchini, A.; Piletsky, S. CHAPTER 1:Nano-Sized Molecularly Imprinted Polymers as Artificial Antibodies. In Molecularly Imprinted Polymers for Analytical Chemistry Applications; Royal Society of Chemistry: London, UK, 2018; pp. 1–27. [Google Scholar]
- Kelley, B. Industrialization of MAb Production Technology: The Bioprocessing Industry at a Crossroads. mAbs 2009, 1, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Saylan, Y.; Denizli, A. Molecular Fingerprints of Hemoglobin on a Nanofilm Chip. Sensors 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Kupai, J.; Razali, M.; Buyuktiryaki, S.; Kecili, R.; Szekely, G. Long-Term Stability and Reusability of Molecularly Imprinted Polymers. Polym. Chem. 2017, 8, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Linares, A.V.; Falcimaigne-Cordin, A.; Gheber, L.A.; Haupt, K. Patterning Nanostructured, Synthetic, Polymeric Receptors by Simultaneous Projection Photolithography, Nanomolding, and Molecular Imprinting. Small Weinh. Bergstr. Ger. 2011, 7, 2318–2325. [Google Scholar] [CrossRef] [PubMed]
- Osman, B.; Uzun, L.; Beşirli, N.; Denizli, A. Microcontact Imprinted Surface Plasmon Resonance Sensor for Myoglobin Detection. Mater. Sci. Eng. C Mater. Biol. Appl. 2013, 33, 3609–3614. [Google Scholar] [CrossRef] [PubMed]
- Yola, M.L.; Atar, N. Development of Cardiac Troponin-I Biosensor Based on Boron Nitride Quantum Dots Including Molecularly Imprinted Polymer. Biosens. Bioelectron. 2019, 126, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Svenson, J.; Nicholls, I.A. On the Thermal and Chemical Stability of Molecularly Imprinted Polymers. Anal. Chim. Acta 2001, 435, 19–24. [Google Scholar] [CrossRef]
- Yusof, N.A.; Rahman, S.K.A.; Hussein, M.Z.; Ibrahim, N.A. Preparation and Characterization of Molecularly Imprinted Polymer as SPE Sorbent for Melamine Isolation. Polymers 2013, 5, 1215–1228. [Google Scholar] [CrossRef]
- Peacock, W.F.; Diercks, D.; Birkhahn, R.; Singer, A.J.; Hollander, J.E.; Nowak, R.; Safdar, B.; Miller, C.D.; Peberdy, M.; Counselman, F.; et al. Can a Point-of-Care Troponin I Assay Be as Good as a Central Laboratory Assay? A MIDAS Investigation. Ann. Lab. Med. 2016, 36, 405–412. [Google Scholar] [CrossRef]
- Suzuki, K.; Komukai, K.; Nakata, K.; Kang, R.; Oi, Y.; Muto, E.; Kashiwagi, Y.; Tominaga, M.; Miyanaga, S.; Ishikawa, T.; et al. The Usefulness and Limitations of Point-of-Care Cardiac Troponin Measurement in the Emergency Department. Intern. Med. Tokyo Jpn. 2018, 57, 1673–1680. [Google Scholar] [CrossRef] [PubMed]
- Quidel. Quidel Triage Cardiac Package Insert, 97000HS. Available online: https://www.quidel.com/immunoassays/triage-test-kits/triage-cardiac-panel (accessed on 24 May 2019).
- Bhatnagar, D.; Kaur, I.; Kumar, A. Ultrasensitive Cardiac Troponin I Antibody Based Nanohybrid Sensor for Rapid Detection of Human Heart Attack. Int. J. Biol. Macromol. 2017, 95, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Han, C.-H.; Jang, J. Rapid Electrical Immunoassay of the Cardiac Biomarker Troponin I through Dielectrophoretic Concentration Using Imbedded Electrodes. Biosens. Bioelectron. 2016, 82, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Shen, X.-L.; Wang, H.-S.; Tao, J.; Huang, J.-Z.; Zeng, Q.; Wang, L.-S. MIPs-Graphene Nanoplatelets-MWCNTs Modified Glassy Carbon Electrode for the Determination of Cardiac Troponin I. Anal. Biochem. 2017, 520, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, S.; Sadanandan, S.; Haridas, V.; Voelcker, N.H.; Prieto-Simon, B. Novel Peptidylated Surfaces for Interference-Free Electrochemical Detection of Cardiac Troponin I. Biosens. Bioelectron. 2018, 99, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Zhao, X.; Ju, X.; Qiu, S.; Hu, W.; Fan, T.; Zhang, J. A New Molecularly Imprinted Polymer (MIP)-Based Electrochemical Sensor for Monitoring Cardiac Troponin I (CTnI) in the Serum. Electroanalysis 2016, 28, 2044–2049. [Google Scholar] [CrossRef]
- Silva, B.V.M.; Rodríguez, B.A.G.; Sales, G.F.; Sotomayor, M.D.P.T.; Dutra, R.F. An Ultrasensitive Human Cardiac Troponin T Graphene Screen-Printed Electrode Based on Electropolymerized-Molecularly Imprinted Conducting Polymer. Biosens. Bioelectron. 2016, 77, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.V.M.; Cavalcanti, I.T.; Silva, M.M.S.; Dutra, R.F. A Carbon Nanotube Screen-Printed Electrode for Label-Free Detection of the Human Cardiac Troponin T. Talanta 2013, 117, 431–437. [Google Scholar] [CrossRef]
- Karimian, N.; Vagin, M.; Zavar, M.H.A.; Chamsaz, M.; Turner, A.P.F.; Tiwari, A. An Ultrasensitive Molecularly-Imprinted Human Cardiac Troponin Sensor. Biosens. Bioelectron. 2013, 50, 492–498. [Google Scholar] [CrossRef]
- Gomes-Filho, S.L.R.; Dias, A.C.M.S.; Silva, M.M.S.; Silva, B.V.M.; Dutra, R.F. A Carbon Nanotube-Based Electrochemical Immunosensor for Cardiac Troponin T. Microchem. J. 2013, 109, 10–15. [Google Scholar] [CrossRef]
- Sharma, D.; Lee, J.; Shin, H. An Electrochemical Immunosensor Based on a 3D Carbon System Consisting of a Suspended Mesh and Substrate-Bound Interdigitated Array Nanoelectrodes for Sensitive Cardiac Biomarker Detection. Biosens. Bioelectron. 2018, 107, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, S.K.; Chen, R.; Kukkar, M.; Song, C.K.; Mutreja, R.; Singh, S.; Paul, A.K.; Lee, H.; Kim, K.-H.; Deep, A.; et al. A Label-Free Electrochemical Immunosensor for the Detection of Cardiac Marker Using Graphene Quantum Dots (GQDs). Biosens. Bioelectron. 2016, 86, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhang, D.; Cui, H.; Huang, T. ZnO/Porous Carbon Composite from a Mixed-Ligand MOF for Ultrasensitive Electrochemical Immunosensing of C-Reactive Protein. Sens. Actuators B Chem. 2019, 284, 354–361. [Google Scholar] [CrossRef]
- Kumar, D.; Prasad, B.B. Multiwalled Carbon Nanotubes Embedded Molecularly Imprinted Polymer-Modified Screen Printed Carbon Electrode for the Quantitative Analysis of C-Reactive Protein. Sens. Actuators B Chem. 2012, 171–172, 1141–1150. [Google Scholar] [CrossRef]
- Bryan, T.; Luo, X.; Bueno, P.R.; Davis, J.J. An Optimised Electrochemical Biosensor for the Label-Free Detection of C-Reactive Protein in Blood. Biosens. Bioelectron. 2013, 39, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Ali, M.A.; Ra, P.; Sharma, A.; Malhotra, B.D.; John, R. Microporous Nanocomposite Enabled Microfluidic Biochip for Cardiac Biomarker Detection. Acs Appl. Mater. Interfaces 2017, 9, 33576–33588. [Google Scholar] [CrossRef]
- Noel, J.; Teizer, W.; Hwang, W. Antifouling Self-Assembled Monolayers on Microelectrodes for Patterning Biomolecules. J. Vis. Exp. JoVE 2009, 30. [Google Scholar] [CrossRef]
- Nogues, C.; Leh, H.; Lautru, J.; Delelis, O.; Buckle, M. Efficient Antifouling Surface for Quantitative Surface Plasmon Resonance Based Biosensor Analysis. PLoS ONE 2012, 7, e44287. [Google Scholar] [CrossRef]
- Campuzano, S.; Pedrero, M.; Yáñez-Sedeño, P.; Pingarrón, J.M. Antifouling (Bio)Materials for Electrochemical (Bio)Sensing. Int. J. Mol. Sci. 2019, 20, 423. [Google Scholar] [CrossRef]
- Wu, J.-G.; Wei, S.-C.; Chen, Y.; Chen, J.-H.; Luo, S.-C. Critical Study of the Recognition between C-Reactive Protein and Surface-Immobilized Phosphorylcholine by Quartz Crystal Microbalance with Dissipation. Langmuir 2018, 34, 943–951. [Google Scholar] [CrossRef]
- Wang, Q.; Jin, H.; Xia, D.; Shao, H.; Peng, K.; Liu, X.; Huang, H.; Zhang, Q.; Guo, J.; Wang, Y.; et al. Biomimetic Polymer-Based Method for Selective Capture of C-Reactive Protein in Biological Fluids. ACS Appl. Mater. Interfaces 2018, 10, 41999–42008. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Kawasaki, H.; Iwasaki, Y. Label-Free Specific Detection and Collection of C-Reactive Protein Using Zwitterionic Phosphorylcholine-Polymer-Protected Magnetic Nanoparticles. Langmuir 2019, 35, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.; Stack, E.; Krivelo, S.; Breheny, M.; Ma, H.; O’Kennedy, R. Enhancing Recombinant Antibody Performance by Optimally Engineering Its Format. J. Immunol. Methods 2018, 463, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Landry, J.P.; Ke, Y.; Yu, G.-L.; Zhu, X.D. Measuring Affinity Constants of 1,450 Monoclonal Antibodies to Peptide Targets with a Microarray-Based Label-Free Assay Platform. J. Immunol. Methods 2015, 417, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.; Hsu, H.-Y.; Wu, H.-T.; Tseng, K.-Y.; Chiou, A.; Yu, C.-J.; Lee, Z.-Y.; Chan, T.-S. Fiber Optic Biosensor for the Detection of C-Reactive Protein and the Study of Protein Binding Kinetics. J. Biomed. Opt. 2007, 12, 024025. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.H.F.; Hartmann, M.; Keusgen, M. SPR-Based Immunosensor for the CRP Detection--a New Method to Detect a Well Known Protein. Biosens. Bioelectron. 2006, 21, 1987–1990. [Google Scholar] [CrossRef] [PubMed]
- Karimian, N.; Turner, A.P.F.; Tiwari, A. Electrochemical Evaluation of Troponin T Imprinted Polymer Receptor. Biosens. Bioelectron. 2014, 59, 160–165. [Google Scholar] [CrossRef]
- Keçili, R. Selective Recognition of Myoglobin in Biological Samples Using Molecularly Imprinted Polymer-Based Affinity Traps. Int. J. Anal. Chem. 2018, 2018, 9. [Google Scholar]
- Cenci, L.; Anesi, A.; Busato, M.; Guella, G.; Bossi, A.M. Molecularly Imprinted Polymers Coupled to Matrix Assisted Laser Desorption Ionization Mass Spectrometry for Femtomoles Detection of Cardiac Troponin I Peptides. J. Mol. Recognit. 2016, 29, 41–50. [Google Scholar] [CrossRef]
- Patel, M.; Dunford, J.V.; Aguilar, S.; Castillo, E.; Patel, E.; Fisher, R.; Ochs, G.; Mahmud, E. Pre-Hospital Electrocardiography by Emergency Medical Personnel: Effects on Scene and Transport Times for Chest Pain and ST-Segment Elevation Myocardial Infarction Patients. J. Am. Coll. Cardiol. 2012, 60, 806–811. [Google Scholar] [CrossRef][Green Version]
- Yaghi, S.; Chang, A.D.; Ricci, B.A.; Jayaraman, M.V.; McTaggart, R.A.; Hemendinger, M.; Narwal, P.; Dakay, K.; Mac Grory, B.; Cutting, S.M.; et al. Early Elevated Troponin Levels After Ischemic Stroke Suggests a Cardioembolic Source. Stroke 2018, 49, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Kanjwal, K.; Imran, N.; Grubb, B.; Kanjwal, Y. Troponin Elevation in Patients with Various Tachycardias and Normal Epicardial Coronaries. Indian Pacing Electrophysiol. J. 2008, 8, 172–174. [Google Scholar] [PubMed]
- Costabel, J.P.; Burgos, L.M.; Trivi, M. The Significance Of Troponin Elevation In Atrial Fibrillation. J. Atr. Fibrillation 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Agewall, S.; Giannitsis, E.; Jernberg, T.; Katus, H. Troponin Elevation in Coronary vs. Non-Coronary Disease. Eur. Heart J. 2011, 32, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Twerenbold, R.; Boeddinghaus, J.; Nestelberger, T.; Wildi, K.; Rubini Gimenez, M.; Badertscher, P.; Mueller, C. Clinical Use of High-Sensitivity Cardiac Troponin in Patients With Suspected Myocardial Infarction. J. Am. Coll. Cardiol. 2017, 70, 996–1012. [Google Scholar] [CrossRef] [PubMed]
- Roffi, M.; Patrono, C.; Collet, J.-P.; Mueller, C.; Valgimigli, M.; Andreotti, F.; Bax, J.J.; Borger, M.A.; Brotons, C.; Chew, D.P.; et al. 2015 ESC Guidelines for the Management of Acute Coronary Syndromes in Patients Presenting without Persistent ST-Segment Elevation: Task Force for the Management of Acute Coronary Syndromes in Patients Presenting without Persistent ST-Segment Elevation of the European Society of Cardiology (ESC). Eur. Heart J. 2016, 37, 267–315. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.; Biener, M.; Vafaie, M.; Doerr, S.; Keller, T.; Blankenberg, S.; Katus, H.A.; Giannitsis, E. Absolute and Relative Kinetic Changes of High-Sensitivity Cardiac Troponin T in Acute Coronary Syndrome and in Patients with Increased Troponin in the Absence of Acute Coronary Syndrome. Clin. Chem. 2012, 58, 209–218. [Google Scholar] [CrossRef]
- Chapman, A.R.; Fujisawa, T.; Lee, K.K.; Andrews, J.P.; Anand, A.; Sandeman, D.; Ferry, A.V.; Stewart, S.; Marshall, L.; Strachan, F.E.; et al. Novel High-Sensitivity Cardiac Troponin I Assay in Patients with Suspected Acute Coronary Syndrome. Heart 2019, 105, 616–622. [Google Scholar] [CrossRef]
- Reichlin, T.; Irfan, A.; Twerenbold, R.; Reiter, M.; Hochholzer, W.; Burkhalter, H.; Bassetti, S.; Steuer, S.; Winkler, K.; Peter, F.; et al. Utility of Absolute and Relative Changes in Cardiac Troponin Concentrations in the Early Diagnosis of Acute Myocardial Infarction. Circulation 2011, 124, 136–145. [Google Scholar] [CrossRef]
- Ambavane, A.; Lindahl, B.; Giannitsis, E.; Roiz, J.; Mendivil, J.; Frankenstein, L.; Body, R.; Christ, M.; Bingisser, R.; Alquezar, A.; et al. Economic Evaluation of the One-Hour Rule-out and Rule-in Algorithm for Acute Myocardial Infarction Using the High-Sensitivity Cardiac Troponin T Assay in the Emergency Department. PLoS ONE 2017, 12, e0187662. [Google Scholar] [CrossRef]
- Kemper, D.W.; Semjonow, V.; de Theije, F.; Keizer, D.; van Lippen, L.; Mair, J.; Wille, B.; Christ, M.; Geier, F.; Hausfater, P.; et al. Analytical Evaluation of a New Point of Care System for Measuring Cardiac Troponin I. Clin. Biochem. 2017, 50, 174–180. [Google Scholar] [CrossRef] [PubMed]
- 231. Greenland, P.; Alpert, J.S.; Beller, G.A.; Benjamin, E.J.; Budoff, M.J.; Fayad, Z.A.; Foster, E.; Hlatky, M.A.; Hodgson, J.M.; Kushner, F.G.; et al. 2010 ACCF/AHA Guideline for Assessment of Cardiovascular Risk in Asymptomatic Adults: Executive Summary. Circulation 2010, 122, 2748–2764. [Google Scholar] [CrossRef]
- Björck, L.; Nielsen, S.; Jernberg, T.; Zverkova-Sandström, T.; Giang, K.W.; Rosengren, A. Absence of Chest Pain and Long-Term Mortality in Patients with Acute Myocardial Infarction. Open Heart 2018, 5, e000909. [Google Scholar] [CrossRef] [PubMed]
- Sara, J.D.; Eleid, M.F.; Gulati, R.; Holmes, D.R. Sudden Cardiac Death from the Perspective of Coronary Artery Disease. Mayo Clin. Proc. 2014, 89, 1685–1698. [Google Scholar] [CrossRef] [PubMed]
- Zellweger, M.J.; Haaf, P.; Maraun, M.; Osterhues, H.H.; Keller, U.; Müller-Brand, J.; Jeger, R.; Pfister, O.; Brinkert, M.; Burkard, T.; et al. Predictors and Prognostic Impact of Silent Coronary Artery Disease in Asymptomatic High-Risk Patients with Diabetes Mellitus. Int. J. Cardiol. 2017, 244, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Piepoli, M.F.; Hoes, A.W.; Agewall, S.; Albus, C.; Brotons, C.; Catapano, A.L.; Cooney, M.-T.; Corrà, U.; Cosyns, B.; Deaton, C.; et al. 2016 European Guidelines on Cardiovascular Disease Prevention in Clinical Practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (Constituted by Representatives of 10 Societies and by Invited Experts)Developed with the Special Contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR). Eur. Heart J. 2016, 37, 2315–2381. [Google Scholar] [CrossRef]
- Cooney, M.T.; Dudina, A.; D’Agostino, R.; Graham, I.M. Cardiovascular Risk-Estimation Systems in Primary Prevention. Circulation 2010, 122, 300–310. [Google Scholar] [CrossRef]
- Chunta, S.; Suedee, R.; Lieberzeit, P.A. High-Density Lipoprotein Sensor Based on Molecularly Imprinted Polymer. Anal. Bioanal. Chem. 2018, 410, 875–883. [Google Scholar] [CrossRef]
- Chunta, S.; Suedee, R.; Lieberzeit, P.A. Low-Density Lipoprotein Sensor Based on Molecularly Imprinted Polymer. Anal. Chem. 2016, 88, 1419–1425. [Google Scholar] [CrossRef]
- Assmann, G.; Schulte, H.; von Eckardstein, A.; Huang, Y. High-Density Lipoprotein Cholesterol as a Predictor of Coronary Heart Disease Risk. The PROCAM Experience and Pathophysiological Implications for Reverse Cholesterol Transport. Atherosclerosis 1996, 124, S11–S20. [Google Scholar] [CrossRef]
- Corti, M.C.; Guralnik, J.M.; Salive, M.E.; Harris, T.; Field, T.S.; Wallace, R.B.; Berkman, L.F.; Seeman, T.E.; Glynn, R.J.; Hennekens, C.H. HDL Cholesterol Predicts Coronary Heart Disease Mortality in Older Persons. JAMA 1995, 274, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Resnick, H.E.; Jablonski, K.A.; Jones, K.L.; Jain, A.K.; Howard, W.J.; Robbins, D.C.; Howard, B.V. Non-HDL Cholesterol as a Predictor of Cardiovascular Disease in Type 2 Diabetes: The Strong Heart Study. Diabetes Care 2003, 26, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Castelli, W.P.; Garrison, R.J.; Wilson, P.W.; Abbott, R.D.; Kalousdian, S.; Kannel, W.B. Incidence of Coronary Heart Disease and Lipoprotein Cholesterol Levels. The Framingham Study. JAMA 1986, 256, 2835–2838. [Google Scholar] [CrossRef] [PubMed]
- Sacks, F.M. The Role of High-Density Lipoprotein (HDL) Cholesterol in the Prevention and Treatment of Coronary Heart Disease: Expert Group Recommendations. Am. J. Cardiol. 2002, 90, 139–143. [Google Scholar] [CrossRef]
- Staněk, V.; Gebauerová, M.; Piťha, J.; Poledne, R.; Lánská, V.; Cífková, R.; Mrázková, J.; Kettner, J. The Risk Profile of Patients with Acute Coronary Syndrome Treated at IKEM between 2006 and 2013. Cor Vasa 2017, 59, e119–e127. [Google Scholar] [CrossRef]
- Jalali, M.T.; Honomaror, A.M.; Rekabi, A.; Latifi, M. Reference Ranges for Serum Total Cholesterol, HDL-Cholesterol, LDL-Cholesterol, and VLDL-Cholesterol and Triglycerides in Healthy Iranian Ahvaz Population. Indian J. Clin. Biochem. 2013, 28, 277–282. [Google Scholar] [CrossRef][Green Version]
- Kaufman, H.W.; Blatt, A.J.; Huang, X.; Odeh, M.A.; Superko, H.R. Blood Cholesterol Trends 2001–2011 in the United States: Analysis of 105 Million Patient Records. PLoS ONE 2013, 8, e63416. [Google Scholar] [CrossRef]
- Gulayin, P.; Irazola, V.; Lozada, A.; Chaparro, M.; Santero, M.; Gutierrez, L.; Poggio, R.; Beratarrechea, A.; Rubinstein, A. Educational Intervention to Improve Effectiveness in Treatment and Control of Patients with High Cardiovascular Risk in Low-Resource Settings in Argentina: Study Protocol of a Cluster Randomised Controlled Trial. BMJ Open 2017, 7, e014420. [Google Scholar] [CrossRef]
- Superko, H.R.; Williams, P.T.; Dansinger, M.; Schaefer, E. Trends in Low-Density Lipoprotein-Cholesterol Blood Values between 2012 and 2017 Suggest Sluggish Adoption of the Recent 2013 Treatment Guidelines. Clin. Cardiol. 2019, 42, 101–110. [Google Scholar] [CrossRef]
- Assessing National Capacity for the Prevention and Control of Noncommunicable Diseases: Report of the 2017 Global Survey; World Health Organization: Geneva, Switzerland, 2018.
- Gupta, M.; Singh, N.; Tsigoulis, M.; Kajil, M.; Hirjikaka, S.; Quan, A.; Teoh, H.; Verma, S. Perceptions of Canadian Primary Care Physicians Towards Cardiovascular Risk Assessment and Lipid Management. Can. J. Cardiol. 2012, 28, 14–19. [Google Scholar] [CrossRef]
- Berger, J.S.; Jordan, C.O.; Lloyd-Jones, D.; Blumenthal, R.S. Screening for Cardiovascular Risk in Asymptomatic Patients. J. Am. Coll. Cardiol. 2010, 55, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Koenig, W. C-Reactive Protein and Cardiovascular Risk: Has the Time Come for Screening the General Population? Clin. Chem. 2001, 47, 9–10. [Google Scholar] [PubMed]
- Myers, G.L.; Christenson, R.H.M.; Cushman, M.; Ballantyne, C.M.; Cooper, G.R.; Pfeiffer, C.M.; Grundy, S.M.; Labarthe, D.R.; Levy, D.; Rifai, N.; et al. National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines: Emerging Biomarkers for Primary Prevention of Cardiovascular Disease. Clin. Chem. 2009, 55, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Jellinger, P.; Smith, D.; Mehta, A.; Ganda, O.; Handelsman, Y.; Rodbard, H.; Shepherd, M.; Seibel, J. American Association of Clinical Endocrinologists’ Guidelines for Management of Dyslipidemia and Prevention of Atherosclerosis. Endocr. Pract. 2012, 18 (Suppl. 1), 1–78. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.-C.; Chen, C.-P.; Horng, J.-C.; Chen, S.-Y. Point-of-Care Protein Sensing Platform Based on Immuno-like Membrane with Molecularly-Aligned Nanocavities. Biosens. Bioelectron. 2013, 50, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Byrne, H.; O’Kennedy, R.J. Antibodies and Antibody-Derived Analytical Biosensors. In Biosensor Technologies for Detection of Biomolecules; Estrela, P., Ed.; Portland Press Ltd.: London, UK, 2016; pp. 9–18. [Google Scholar]
- Karmali, K.N.; Brown, T.; Sanchez, T.; Long, T.; Persell, S.D. Point-of-Care Testing to Promote Cardiovascular Disease Risk Assessment: A Proof of Concept Study. Prev. Med. Rep. 2017, 7, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Haggerty, L.; Tran, D. Cholesterol Point-of-Care Testing for Community Pharmacies: A Review of the Current Literature. J. Pharm. Pract. 2017, 30, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Dale, R.A.; Jensen, L.H.; Krantz, M.J. Comparison of Two Point-of-Care Lipid Analyzers for Use in Global Cardiovascular Risk Assessments. Ann. Pharmacother. 2008, 42, 633–639. [Google Scholar] [CrossRef]
- Whitehead, S.J.; Ford, C.; Gama, R. The Impact of Different Point-of-Care Testing Lipid Analysers on Cardiovascular Disease Risk Assessment. J. Clin. Pathol. 2014, 67, 535–539. [Google Scholar] [CrossRef]
- Bubner, T.K.; Laurence, C.O.; Gialamas, A.; Yelland, L.N.; Ryan, P.; Willson, K.J.; Tideman, P.; Worley, P.; Beilby, J.J. Effectiveness of Point-of-Care Testing for Therapeutic Control of Chronic Conditions: Results from the PoCT in General Practice Trial. Med. J. Aust. 2009, 190, 624–626. [Google Scholar]




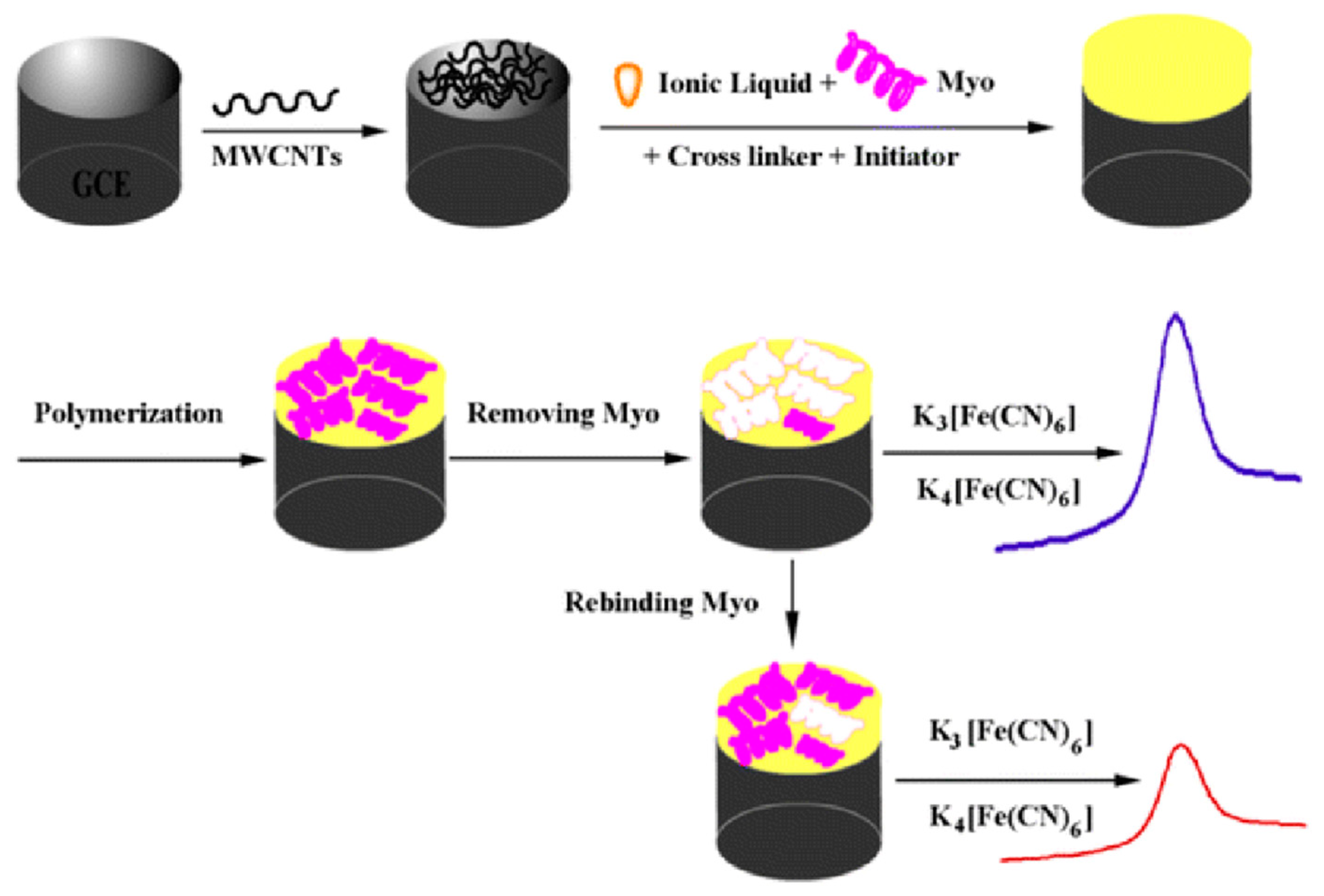
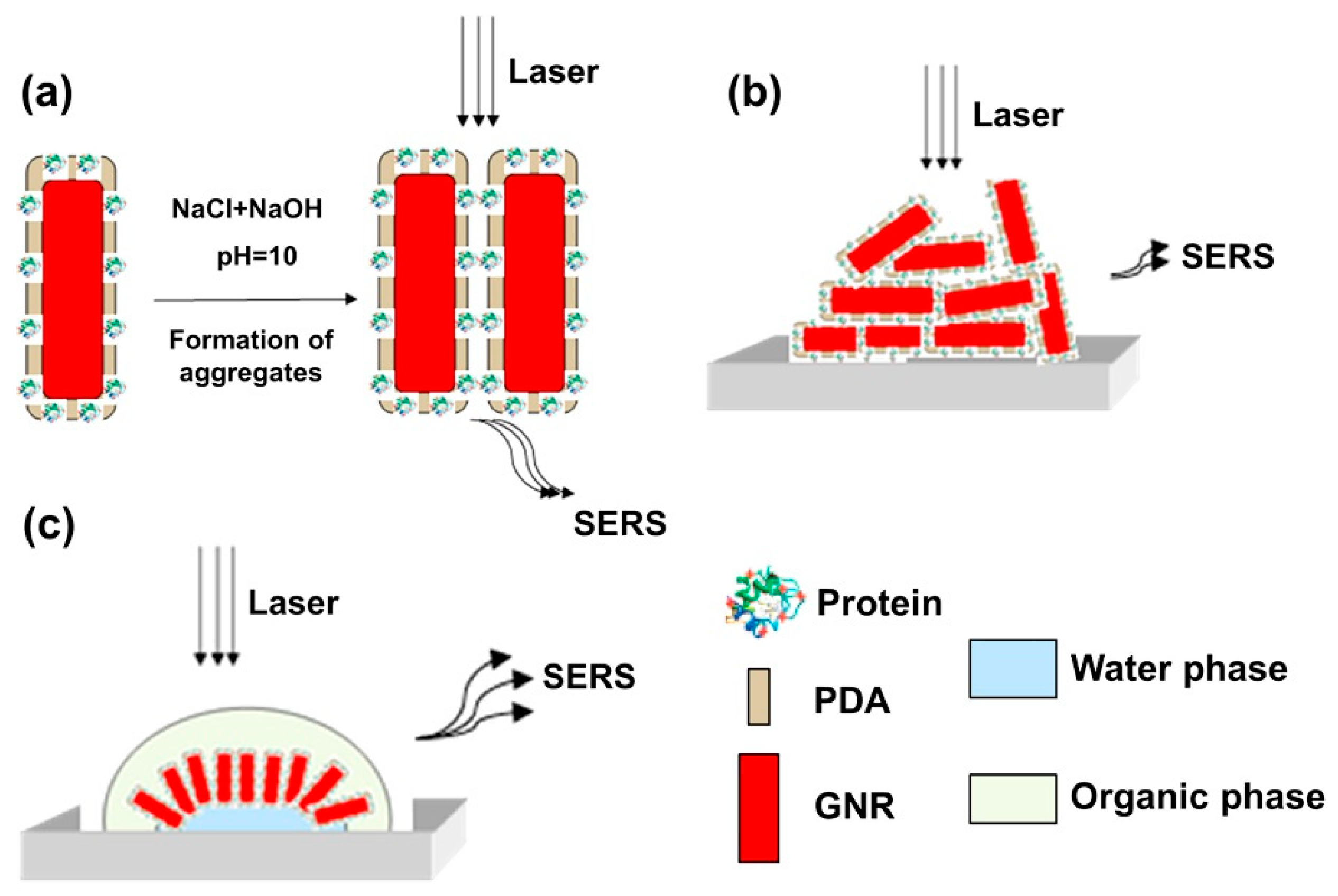




| Challenges of PIP Synthesis | Potential PIP Deficiencies | Significance for Sensor Performance | |
|---|---|---|---|
| Description | Illustrations | ||
| Template/ Monomer/ Cross-linker Ratio | Excessive cross-linker can block access to binding sites, therefore restricting mass transfer |  | The time period required to perform the measurements is increased |
| Suboptimal mixture can result in a PIP with a lack of binding sites |  | Limited detection range | |
| A low cross-linker ratio can lead to binding sites lacking rigidity | 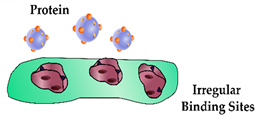 | Recognition element with low specificity and an increase in non-specific adsorption | |
| Excessive cross-linker can lead to template entrapment |  | Inability to rebind target analyte due to binding sites being occupied/ inaccessible | |
| Monomer/ Cross-linker/ Solvent Selection | Solvent selection is typically limited to aqueous-based to ensure proteins retain conformational structure |  | Possible production of imprinted polymers with poor performance characteristics |
| Monomer and cross-linker selection restricted to those soluble in aqueous-based solvents |  | ||
| Template Extraction | Harsh extraction solvents can damage binding sites and reduce binding site homogeneity |  | Heterogeneous binding sites have poor specificity and can also affect binding kinetics |
| Incomplete template removal can produce template leaching during measurements |  | Release of template protein while performing measurements can induce a false response | |
| Suboptimal extraction technique can result in high affinity binding sites retaining the template protein |  | Only low affinity binding sites remain accessible, thus, the PIP will have low specificity towards the template | |
| Enzymatic digestion can partially remove the template protein from binding sites |  | Occupancy of binding sites can inhibit rebinding and therefore limit the detection range | |
| Sensor Type | Target Biomarker | LOD (ng/L) | Linear Range (ng/mL) | Response Time * (min) | Reference |
|---|---|---|---|---|---|
| Biosensor | cTnI | 0.025 | 0.000166–16.5 | 10 | [191] |
| Biomimetic | cTnI | 0.5 | 0.01–5.00 | ND | [185] |
| Biosensor | cTnI | 0.7 | 0.001–100 | 1 | [192] |
| Biomimetic | cTnI | 0.8 | 0.005–60 | 10 | [193] |
| Biosensor | cTnI | 1.9 | 0.00001–1.0 | 30 | [194] |
| Biomimetic | cTnI | 634.5 | 1.18–117.5 | 5 | [195] |
| Biomimetic | cTnT | 6 | 0.01–0.1 | 30 | [196] |
| Biosensor | cTnT | 7 | 0.005–0.065 | 40 | [197] |
| Biomimetic | cTnT | 9 | 0.009–0.8 | 10 | [198] |
| Biosensor | cTnT | 33 | 0.1–10 | 60 | [199] |
| Biomimetic | MYO | 0.045 | 0.000304–0.571 | 30 | [136] |
| Biosensor | MYO | 0.4 | 0.001–100 | 130 | [200] |
| Biosensor | MYO | 10 | 0.01–100 | 10 | [201] |
| Biomimetic | MYO | 26300 | 100–1000 | 25 | [184] |
| Biosensor | CRP | 5.0 | 0.01–1000 | 60 | [202] |
| Biomimetic | CRP | 500 | 180–8510 | 3 | [203] |
| Biosensor | CRP | 22000 | 62.5–6250 | 30 | [204] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Regan, B.; Boyle, F.; O’Kennedy, R.; Collins, D. Evaluation of Molecularly Imprinted Polymers for Point-of-Care Testing for Cardiovascular Disease. Sensors 2019, 19, 3485. https://doi.org/10.3390/s19163485
Regan B, Boyle F, O’Kennedy R, Collins D. Evaluation of Molecularly Imprinted Polymers for Point-of-Care Testing for Cardiovascular Disease. Sensors. 2019; 19(16):3485. https://doi.org/10.3390/s19163485
Chicago/Turabian StyleRegan, Brian, Fiona Boyle, Richard O’Kennedy, and David Collins. 2019. "Evaluation of Molecularly Imprinted Polymers for Point-of-Care Testing for Cardiovascular Disease" Sensors 19, no. 16: 3485. https://doi.org/10.3390/s19163485
APA StyleRegan, B., Boyle, F., O’Kennedy, R., & Collins, D. (2019). Evaluation of Molecularly Imprinted Polymers for Point-of-Care Testing for Cardiovascular Disease. Sensors, 19(16), 3485. https://doi.org/10.3390/s19163485





