Electrochemical Sensor Based on Poly(Azure B)-DNA Composite for Doxorubicin Determination
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents

2.2. Apparatus
2.3. DNA Sensor Preparation
2.4. Doxorubicin Measurements and Real Sample Assay
3. Results and Discussion
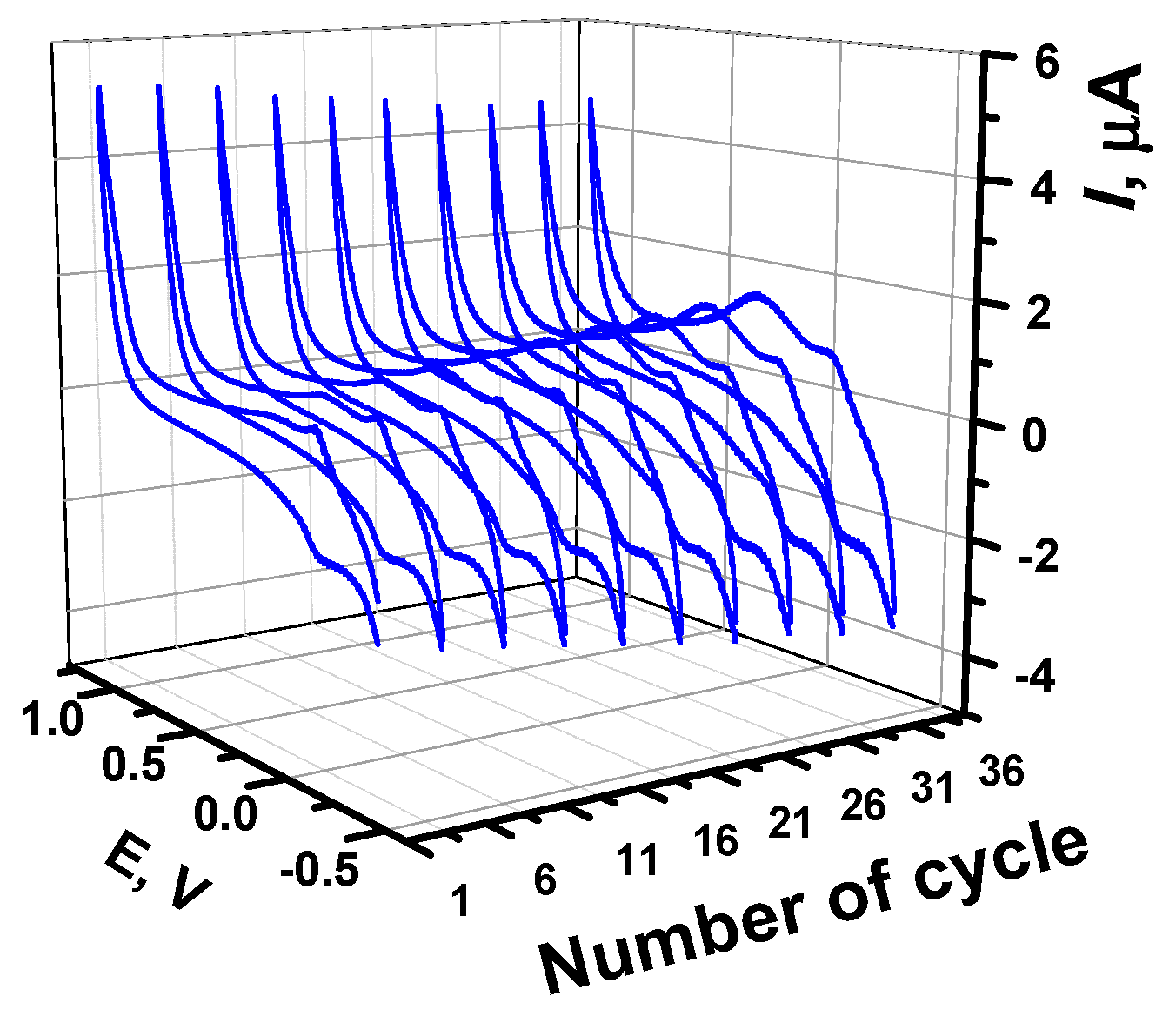
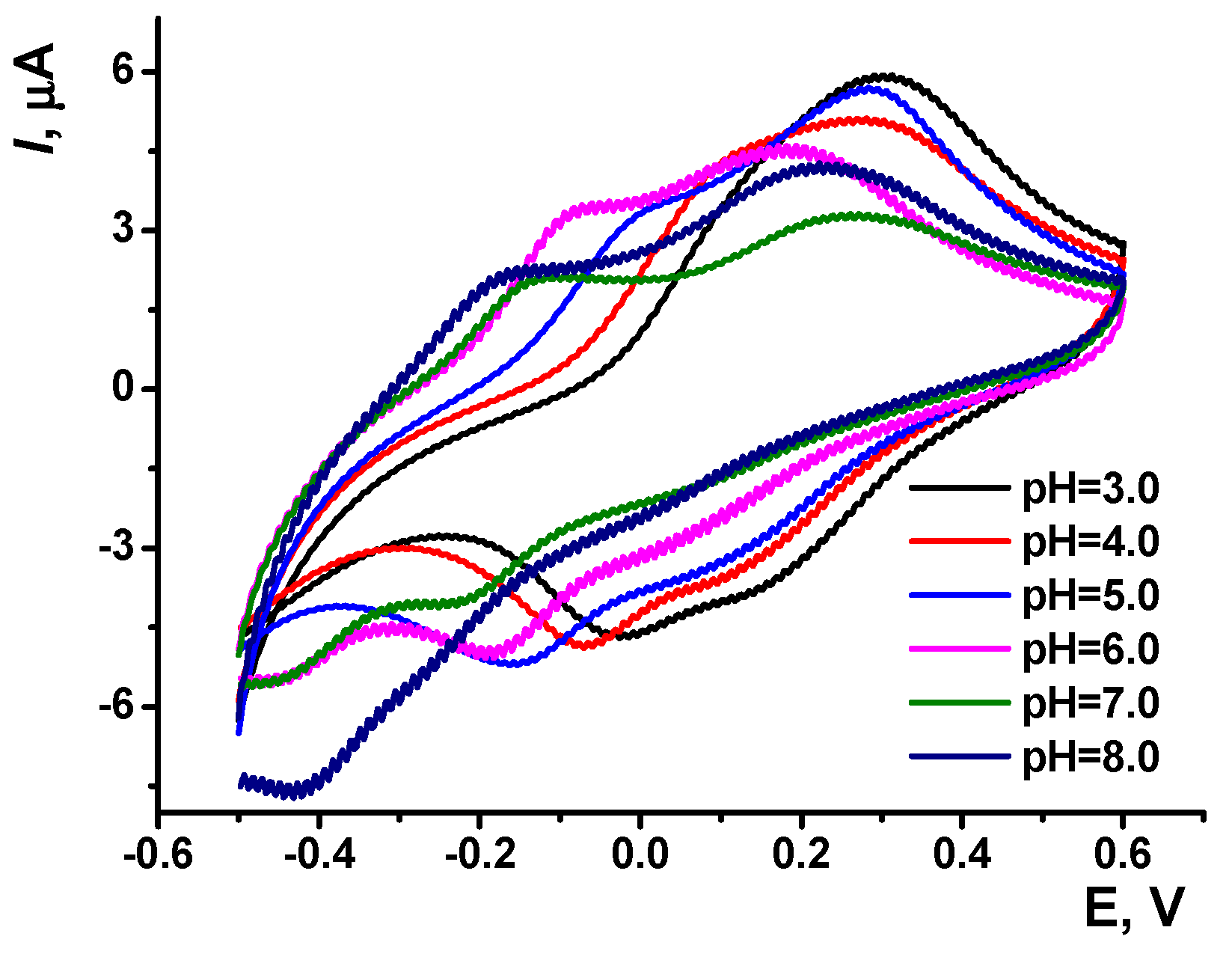
3.1. Azure B Polymerization and Redox Properties of Poly(Azure B) Layer
3.2. DNA Deposition and Determination
0.2–1.0 mg/mL: I/I0, % = (59.6 ± 0.7) − (15.7 ± 0.4) × log(cDNA, mg/mL), R2 = 0.9562, n = 5
3.3. Doxorubicin Determination
3.4. Measurement Precision
3.5. Selectivity and Real Sample Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rahman, Md.M.; Li, X.-B.; Lopa, N.S.; Ahn, S.J.; Lee, J.J. Electrochemical DNA hybridization sensors based on conducting polymers. Sensors 2015, 15, 3801–3829. [Google Scholar] [CrossRef]
- Campuzano, S.; Pedrero, M.; Pingarrón, J.M. Electrochemical nucleic acid-based biosensing of drugs of abuse and pharmaceuticals. Curr. Med. Chem. 2018, 25, 4102–4118. [Google Scholar] [CrossRef] [PubMed]
- Kahanda, D.; Chakrabarti, G.; McWilliams, M.A.; Boothman, D.A.; Slinker, J.D. Using DNA devices to track anticancer drug activity. Biosens. Bioelectron. 2018, 80, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Piedade, J.A.P.; Oliveira, P.S.C.; Lopes, M.C.; Oliveira-Brett, A.M. Voltammetric determination of γ radiation-induced DNA damage. Anal. Biochem. 2006, 355, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Fojta, M.; Daňhel, A.; Havran, L.; Vyskočil, V. Recent progress in electrochemical sensors and assays for DNA damage and repair. TrAC Trends Anal. Chem. 2016, 79, 160–167. [Google Scholar] [CrossRef]
- Fojta, M. Electrochemical Sensors for DNA Interactions and Damage. Electroanalysis 2002, 14, 1449–1463. [Google Scholar] [CrossRef]
- Barroso, M.F.; de-los-Santos-Álvarez, N.; Delerue-Matosa, C.; Oliveira, M.B.P.P. Towards a reliable technology for antioxidant capacity and oxidative damage evaluation: Electrochemical (bio)sensors. Biosens. Bioelectron. 2011, 30, 1–12. [Google Scholar] [CrossRef]
- Palchaudhuri, R.; Hergenrother, P.J. DNA as a target for anticancer compounds: Methods to determine the mode of binding and the mechanism of action. Curr. Opin. Biotechnol. 2007, 18, 497–503. [Google Scholar] [CrossRef]
- Carrion, C.; de Madariaga, M.A.; Domingo, J.C. In vitro cytotoxic study of immunoliposomal doxorubicin targeted to human CD34(+) leukemic cells. Life Sci. 2004, 75, 313–328. [Google Scholar] [CrossRef] [PubMed]
- McGowan, J.V.; Chung, R.; Maulik, A.; Piotrowska, A.; Walker, J.M.; Yellon, D.M. Anthracycline chemotherapy and cardiotoxicity. Cardiovasc. Drugs Ther. 2017, 31, 63–75. [Google Scholar] [CrossRef]
- Agarwal, S.; Jangir, D.K.; Mehrotra, R. Spectroscopic studies of the effects of anticancer drug mitoxantrone interaction with calf-thymus DNA. J. Photochem. Photobiol. B 2013, 120, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Ocak, I.; Kara, H.E.S. Phosphorescent detection of DNA- drug interaction based on emission quenching of ZnS quantum dots via photoinduced electron transfer. J. Lumin. 2018, 197, 112–118. [Google Scholar] [CrossRef]
- Proni, G.; Tami, K.; Berova, N.; Ellestad, G.A. Circular dichroism analysis of the calicheamicin-DNA interaction revisited. J. Pharm. Biomed. Anal. 2017, 144, 1–5. [Google Scholar] [CrossRef]
- Wolf, L.K.; Fullenkamp, D.E.; Georgiadis, R.M. Quantitative angle-resolved SPR imaging of DNA-DNA and DNA-drug kinetics. J. Am. Chem. Soc. 2005, 127, 17453–17459. [Google Scholar] [CrossRef]
- Svitková, V.; Labuda, J. Construction of electrochemical DNA biosensors for investigation of potential risk chemical and physical agents. Monatsh. Chem. 2017, 148, 1569–1579. [Google Scholar] [CrossRef]
- Karimi-Maleh, H.; Bananezhad, A.; Ganjali, M.R.; Norouzi, P.; Sadrni, A. Surface amplification of pencil graphite electrode with polypyrrole and reduced graphene oxide for fabrication of a guanine/adenine DNA based electrochemical biosensors for determination of didanosine anticancer drug. Appl. Surf. Sci. 2018, 441, 55–60. [Google Scholar] [CrossRef]
- Zhao, C.; Jin, H.; Gui, R.; Wang, Z. Facile fabrication of dual-ratiometric electrochemical sensors based on a bare electrode for dual-signal sensing of analytes in electrolyte solution. Sens. Actuators B 2017, 242, 71–78. [Google Scholar] [CrossRef]
- Evtugyn, G.A.; Goldfarb, O.E.; Budnikov, H.C.; Ivanov, A.N.; Vinter, V.G. Amperometric DNA-peroxidase sensor for the detection of pharmaceutical preparations. Sensors 2005, 5, 364–376. [Google Scholar] [CrossRef]
- Sontz, P.A.; Muren, N.B.; Barton, J.K. DNA charge transport for sensing and signaling. Acc. Chem. Res. 2012, 45, 1792–1800. [Google Scholar] [CrossRef]
- Sen, T.; Mishra, S.; Shimpi, N.G. Synthesis and sensing applications of polyaniline nanocomposites: A review. RSC Adv. 2016, 6, 42196–42222. [Google Scholar] [CrossRef]
- Dhand, C.; Das, M.; Datta, M.; Malhotra, B.D. Recent advances in polyaniline based biosensors. Biosens. Bioelectron. 2011, 26, 2811–2821. [Google Scholar] [CrossRef]
- Abedi, M.; Bathaie, S.Z.; Mousavi, M.F. Interaction between DNA and some salicylic acid derivatives and characterization of their DNA targets. Electroanalysis 2013, 25, 2547–2556. [Google Scholar] [CrossRef]
- Paziewska-Nowak, A.; Jankowska-Śliwińska, J.; Dawgul, M.; Pijanowska, D.G. Selective electrochemical detection of pirarubicin by means of DNA-modified graphite biosensor. Electroanalysis 2017, 29, 1810–1819. [Google Scholar] [CrossRef]
- Hajian, R.; Tayebi, Z.; Shams, N. Fabrication of an electrochemical sensor for determination of doxorubicin in human plasma and its interaction with DNA. J. Pharm. Anal. 2017, 7, 27–33. [Google Scholar] [CrossRef]
- Shamagsumova, R.; Porfireva, A.; Stepanova, V.; Osin, Y.; Evtugyn, G.; Hianik, T. Polyaniline–DNA based sensor for the detection of anthracycline drugs. Sens. Actuators B 2015, 220, 573–582. [Google Scholar] [CrossRef]
- Kulikova, T.N.; Porfireva, A.V.; Shamagsumova, R.V.; Evtugyn, G.A. Voltammetric sensor with replaceable polyaniline-DNA layer for doxorubicin determination. Electroanalysis 2018, 30, 2284–2292. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Li, C.; Zeng, X.; Tang, W.; Chen, X. A voltammetric study on the interaction between isoproterenol and cardiomyocyte DNA by using a glassy carbon electrode modified with carbon nanotubes, polyaniline and gold nanoparticles. Microchim. Acta 2017, 184, 2999–3006. [Google Scholar] [CrossRef]
- Meng, L.; Chen, L.; Ge, T.; Yang, T.; Jiao, K. Sulfonated polyaniline-graphene oxide hybrids: Synthesis and effect of monomer composition on the electrochemical signal for direct DNA detection. J. Polym. Sci. A 2016, 54, 1762–1773. [Google Scholar] [CrossRef]
- Porfireva, A.V.; Evtugyn, G.A.; Ivanov, A.N.; Hianik, T. Impedimetric aptasensors based on carbon nanotubes – poly(methylene blue) composite. Electroanalysis 2013, 22, 2187–2195. [Google Scholar] [CrossRef]
- Evtugyn, G.A.; Porfireva, A.V.; Hianik, T.; Cheburova, M.S.; Budnikov, H.C. Potentiometric DNA sensor based on electropolymerized phenothiazines for protein detection. Electroanalysis 2008, 20, 1300–1308. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, Y. Lable-free electrochemical DNA sensor based on gold nanoparticles/poly(neutral red) modified electrode. Electroanalysis 2010, 22, 673–679. [Google Scholar] [CrossRef]
- Kuzin, Y.; Kappo, D.; Porfireva, A.; Shurpik, D.; Stoikov, I.; Evtugyn, G.; Hianik, T. Electrochemical DNA sensor based on carbon black—poly(neutral red) composite for detection of oxidative DNA damage. Sensors 2018, 18, 3489. [Google Scholar] [CrossRef]
- Cai, C.-X.; Xie, K.-H. Electrocatalysis of NADH oxidation with electropolymerized films of azure I. J. Electroanal. Chem. 1997, 427, 147–153. [Google Scholar] [CrossRef]
- Sha, Y.; Gao, Q.; Qi, B.; Yang, X. Electropolymerization of Azure B on a screen-printed carbon electrode and its application to the determination of NADH in a flow injection analysis system. Microchim. Acta 2004, 148, 335–341. [Google Scholar] [CrossRef]
- Kong, Y.; Ou, J.; Liu, Z.; Xue, S.; Tao, Y.; Ma, J. The electrocatalytic characteristics of poly(azure B) and its application in the sensitive determination of hydroquinone. Anal. Methods 2014, 6, 3735–3740. [Google Scholar] [CrossRef]
- Shan, D.; Mousty, C.; Cosnier, S.; Mu, S. A composite poly azure B–clay–enzyme sensor for the mediated electrochemical determination of phenols. J. Electroanal. Chem. 2002, 537, 103–109. [Google Scholar] [CrossRef]
- Bayındır, O.; Alanyalıoğlu, M. Formation mechanism of polymeric thin films of Azure B on gold electrodes. ChemistrySelect 2018, 3, 2167–2173. [Google Scholar] [CrossRef]
- Nishida, Y.; Domura, R.; Sakai, R.; Okamoto, M.; Arakawa, S.; Ishiki, R.; Salick, M.R.; Turng, L.-S. Fabrication of PLLA/HA composite scaffolds modified by DNA. Polymer 2015, 56, 73–81. [Google Scholar] [CrossRef]
- Karyakin, A.A.; Karyakina, E.E.; Schmidt, H.-L. Electropolymerized azines: A new group of electroactive polymers. Electroanalysis 1999, 11, 149–155. [Google Scholar] [CrossRef]
- Guidelli, R.; Compton, R.G.; Feliu, J.M.; Gileadi, E.; Lipkowski, J.; Schmickler, W.; Trasatti, S. Defining the transfer coefficient in electrochemistry: An assessment (IUPAC Technical Report). Pure Appl. Chem. 2014, 86, 245–258. [Google Scholar] [CrossRef]
- Minotti, G.; Menna, P.; Salvatorelli, E.; Cairo, G.; Gianni, L. Anthracyclines molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharm. Rev. 2004, 56, 185–229. [Google Scholar] [CrossRef]
- Carvalho, C.; Santos, R.X.; Cardoso, S.; Correia, S.; Oliveira, P.J.; Santos, M.S.; Moreira, P.I. Doxorubicin: The good, the bad and the ugly effect. Curr. Med. Chem. 2009, 16, 3267–3285. [Google Scholar] [CrossRef]
- Danesi, R.; Fogli, S.; Gennari, A.; Conte, P.; Del Tacca, M. Pharmacokinetic-pharmacodynamic relationships of the anthracycline anticancer drugs. Clin. Pharmacokinet. 2002, 41, 431–444. [Google Scholar] [CrossRef]
- Fahmy, O.T.; Korany, M.A.; Maher, H.M. High performance liquid chromatographic determination of some co-administered anticancer drugs in pharmaceutical preparations and in spiked human plasma. J. Pharm. Biomed. Anal. 2004, 34, 1099–1107. [Google Scholar] [CrossRef]
- Gavenda, A.; Ševčík, J.; Psotová, J.; Bednář, P.; Barták, P.; Adamovský, P.; Šimánek, V. Determination of anthracycline antibiotics doxorubicin and daunorubicin by capillary electrophoresis with UV absorption detection. Electrophoresis 2001, 22, 2782–2785. [Google Scholar] [CrossRef]
- Materon, E.M.; Wong, A.; Fatibello-Filho, O.; Faria, R.C. Development of a simple electrochemical sensor for the simultaneous detection of anticancer drugs. J. Electroanal. Chem. 2018, 827, 64–72. [Google Scholar] [CrossRef]
- Guo, H.; Gui, R.; Wang, Z.; Xia, J.; Zhang, F. Electrodeposition one-step preparation of silver nanoparticles/carbon dots/reduced graphene oxide ternary dendritic nanocomposites for sensitive detection of doxorubicin. Sens. Actuators B. 2017, 253, 50–57. [Google Scholar] [CrossRef]
- Peng, A.; Xu, H.; Luo, C.; Ding, H. Application of a disposable doxorubicin sensor for direct determination of clinical drug concentration in patient blood. Int. J. Electrochem. Sci. 2016, 11, 6266–6278. [Google Scholar] [CrossRef]
- Soleymani, J.; Hasanzadeh, M.; Eskandani, M.; Khoubnasabjafari, M.; Shadjou, N.; Jouyban, A. Electrochemical sensing of doxorubicin in unprocessed whole blood, cell lysate, and human plasma samples using thin film of poly-arginine modified glassy carbon electrode. Mater. Sci. Eng. C 2017, 77, 790–802. [Google Scholar] [CrossRef]
- Alizadeh, P.M.; Hasanzadeh, M.; Soleymani, J.; Gharamaleki, J.V.; Jouyban, A. Application of bioactive cyclic oligosaccharide on the detection of doxorubicin hydrochloride in unprocessed human plasma sample: A new platform towards efficient chemotherapy. Microchem. J. 2019, 145, 450–455. [Google Scholar] [CrossRef]
- Bahner, N.; Reich, P.; Frense, D.; Menger, M.; Schieke, K.; Beckmann, D. An aptamer-based biosensor for detection of doxorubicin by electrochemical impedance spectroscopy. Anal. Bioanal. Chem. 2018, 410, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Evtugyn, G.; Porfireva, A.; Stepanova, V.; Budnikov, H. Electrochemical biosensors based on native DNA and nanosized mediator for the detection of anthracycline preparations. Electroanalysis 2015, 27, 629–637. [Google Scholar] [CrossRef]
- Ceruti, M.; Tagini, V.; Recalenda, V.; Arpicco, S.; Cattel, L.; Airoldi, M.; Bumma, C. Docetaxel in combination with epirubicin in metastatic breast cancer: Pharmacokinetic interactions. Il Farmaco 1999, 54, 733–739. [Google Scholar] [CrossRef]




| Peak Potential, V | log (I, μA) = a + b × log (ν, V/s) | |||
|---|---|---|---|---|
| a | b | R2 | n | |
| 0.25 | 1.08 ± 0.06 | 1.00 ± 0.04 | 0.987 | 8 |
| −0.09 | 0.89 ± 0.02 | 0.90 ± 0.01 | 0.998 | 9 |
| −0.18 | 1.34 ± 0.06 | 1.13 ± 0.07 | 0.983 | 9 |
| Modifier | Concentration Range | LOD, nM | Ref. |
|---|---|---|---|
| Multiwalled carbon nanotubes | 0.09–7.36 µM | 3 | [24] |
| Carbon black, Cu nanoparticles, Nafion | 0.46–5.1 µM | 24 | [46] |
| Ag nanoparticles, carbon dots on reduced graphene oxide | 1.0 µM–10 nM | 2 | [47] |
| Multiwalled carbon nanotubes, poly(lysine) | 2.5 nM–0.25 µM | 1 | [48] |
| Poly(arginine) | 69 nM–1.08 µM | 0.1 | [49] |
| Poly(taurine), β-cyclodextrin and graphene quantum dots | 0.086–3.45 μM | 12 | [50] |
| Aptamer against doxorubicin | 31–125 nM | 28 nM | [51] |
| Polyaniline, DNA | 0.1 nM–0.2 mM | 0.01 | [25] |
| Poly(Neutral red), pillar [5]arene, DNA | 0.01–100 µM | 0.1 | [52] |
| Poly(Azure B) | 0.1 µM–0.3 nM | 0.1 | This work |
| Sample | Media | (I0 – I)/(I0 – Imin), % | Recovery, % |
|---|---|---|---|
| Doxorubicin (Sigma) | Standard solution in HEPES | 78 ± 1 | - |
| + 4 mg/mL BSA | 78 ± 3 | 100 | |
| + 40 mg/mL BSA | 73 ± 5 | 105 | |
| Doxorubicin (Sigma) | Ringer-Locke’s solution | 77 ± 2 | 101 |
| Doxorubicin-LANS ® | Ringer-Locke’s solution | 77 ± 2 | 101 |
| Doxorubicin-TEVA ® | Ringer-Locke’s solution | 80 ± 3 | 97 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porfireva, A.; Vorobev, V.; Babkina, S.; Evtugyn, G. Electrochemical Sensor Based on Poly(Azure B)-DNA Composite for Doxorubicin Determination. Sensors 2019, 19, 2085. https://doi.org/10.3390/s19092085
Porfireva A, Vorobev V, Babkina S, Evtugyn G. Electrochemical Sensor Based on Poly(Azure B)-DNA Composite for Doxorubicin Determination. Sensors. 2019; 19(9):2085. https://doi.org/10.3390/s19092085
Chicago/Turabian StylePorfireva, Anna, Vyatseslav Vorobev, Sofya Babkina, and Gennady Evtugyn. 2019. "Electrochemical Sensor Based on Poly(Azure B)-DNA Composite for Doxorubicin Determination" Sensors 19, no. 9: 2085. https://doi.org/10.3390/s19092085





