Vibrational Spectroscopy as a Sensitive Probe for the Chemistry of Intra-Phase Bacterial Growth
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Growth and Sample Preparation
2.2. Raman Measurements
2.3. Infrared Measurements
2.4. Data Analysis
3. Results and Discussion
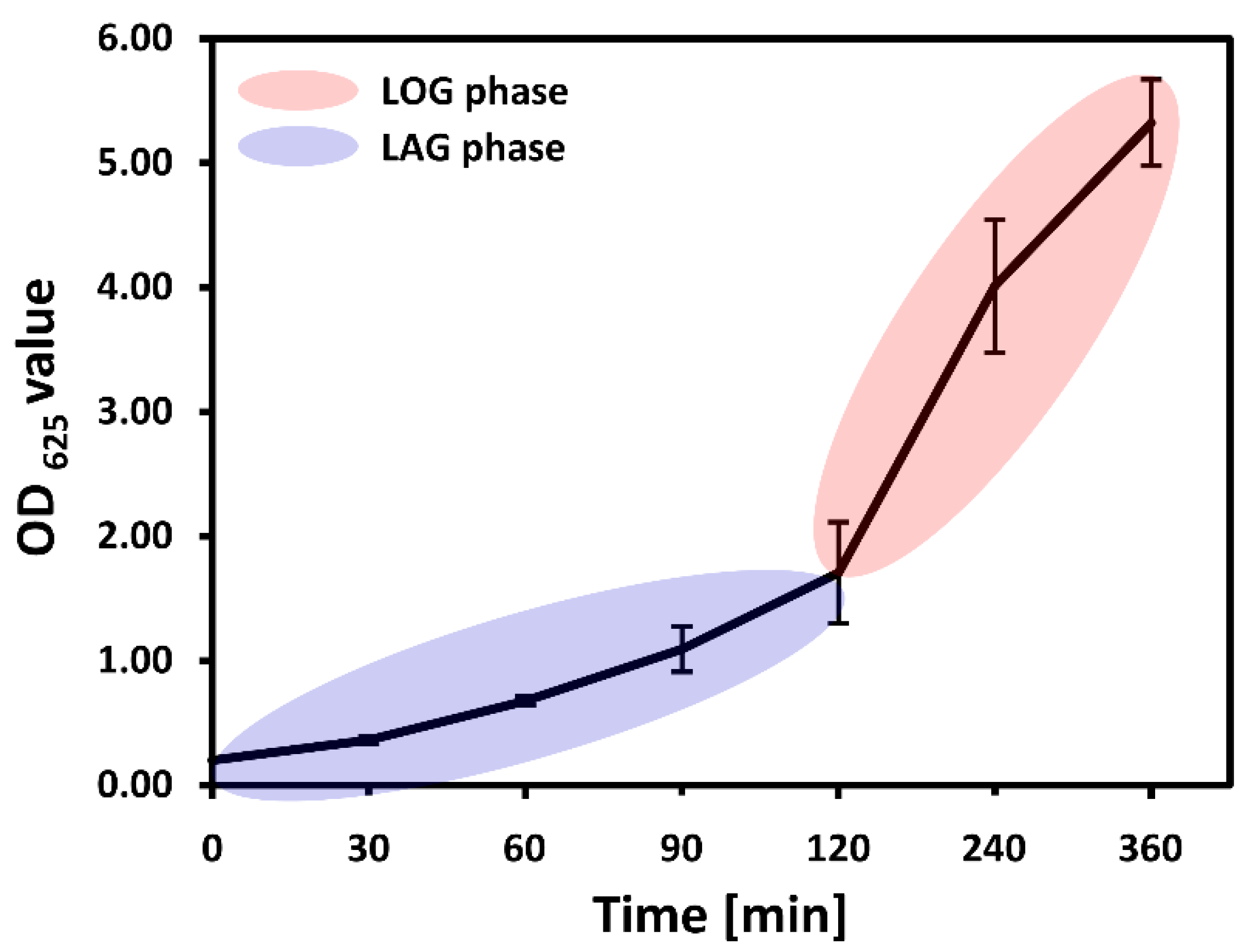
3.1. Growth Phases over the Course of the Experiment
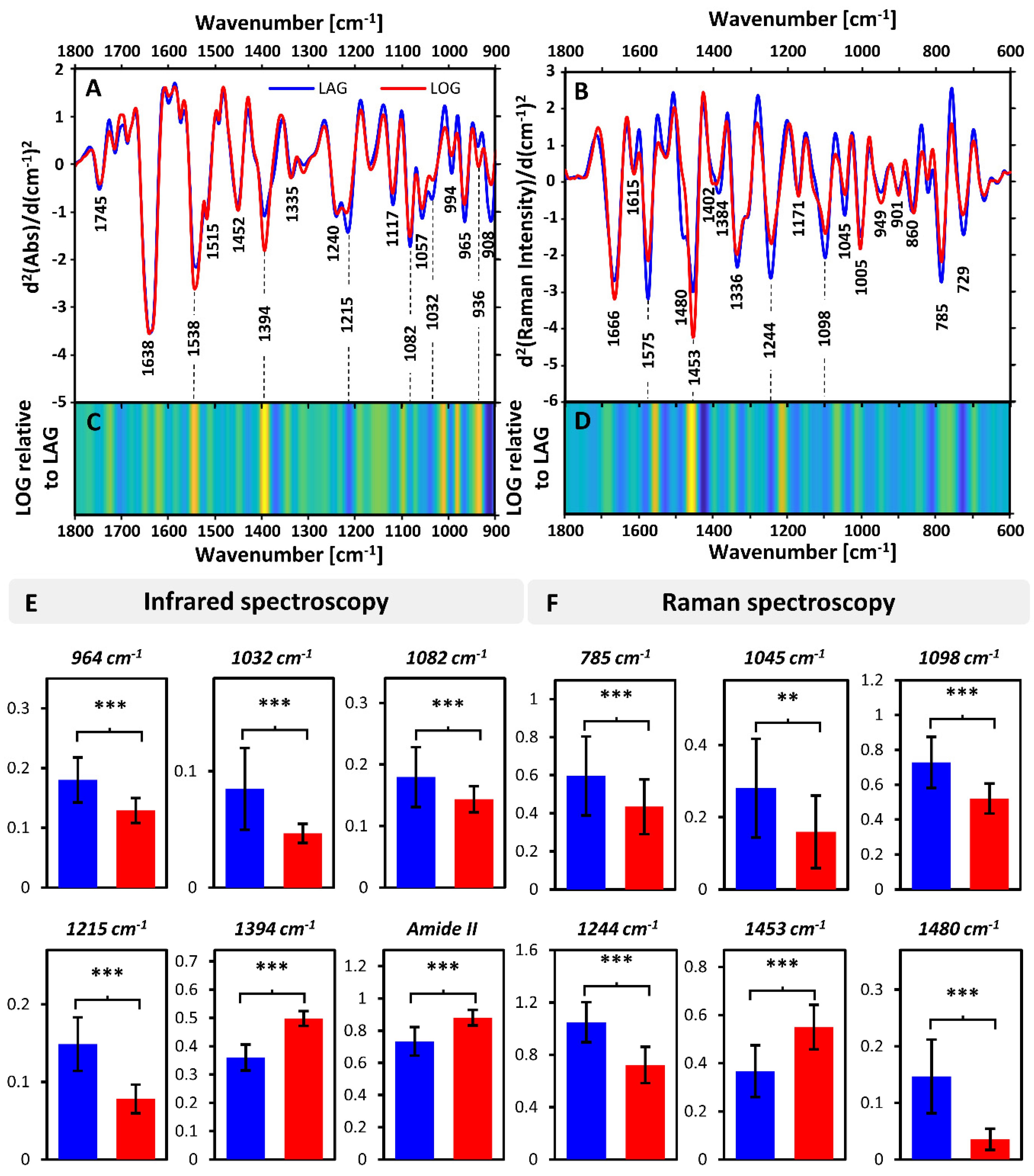
3.2. Inter-Phase Growth Changes
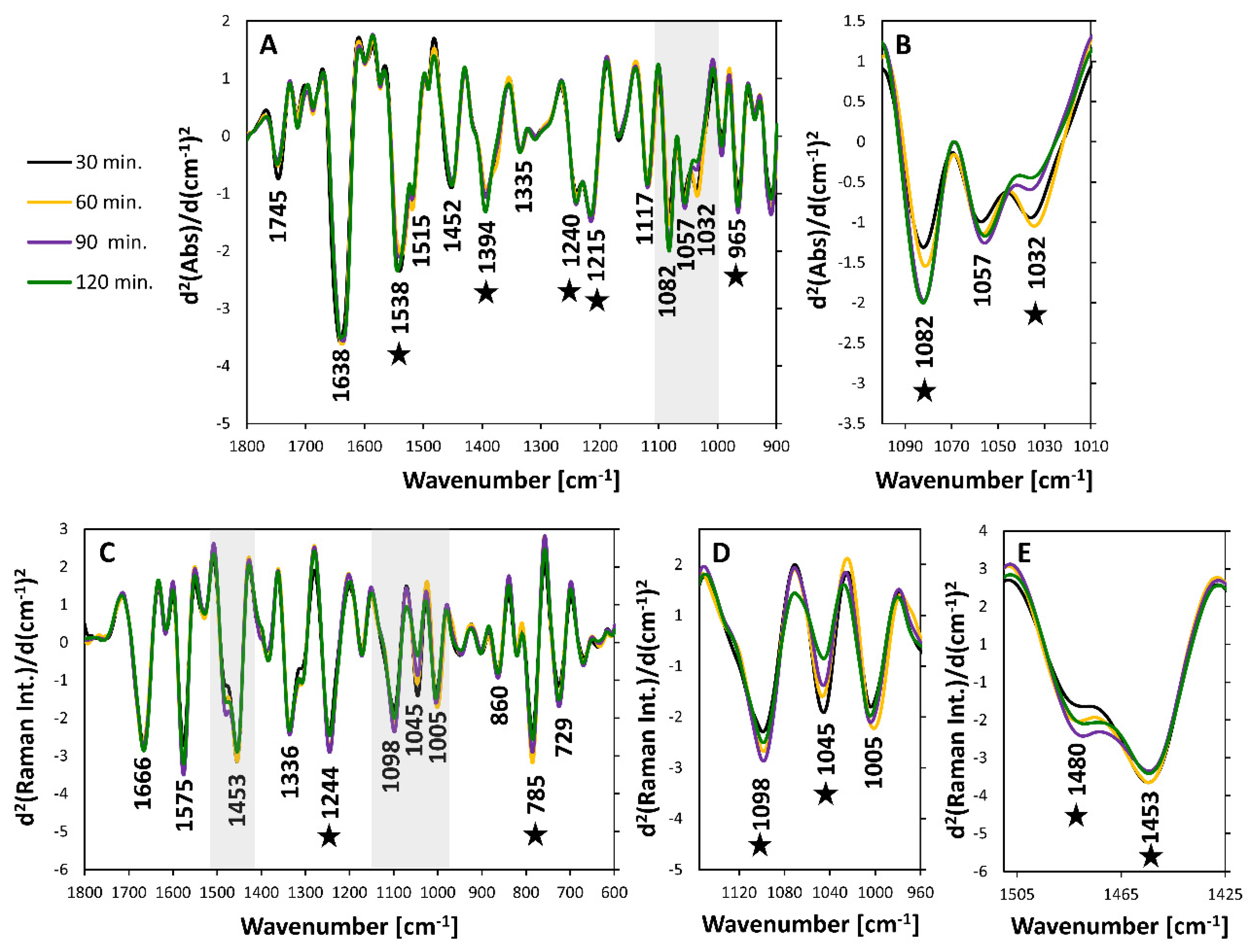
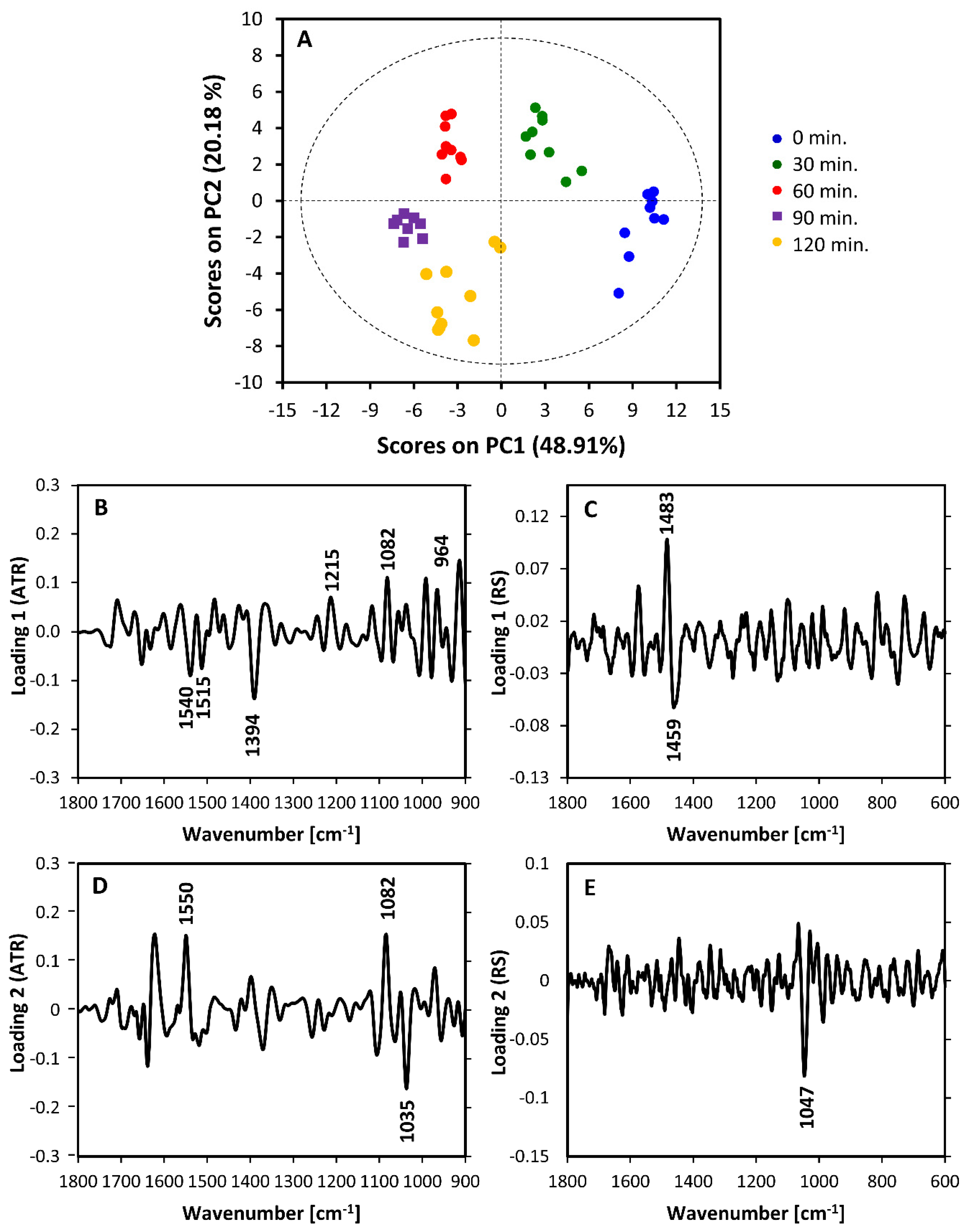
3.3. Intra-Phase Growth Changes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cooper, S. Bacterial Growth and Division: Biochemistry and Regulation of Prokaryotic and Eukaryotic Division Cycles; Elsevier: Amsterdam, The Netherlands, 2012; ISBN 978-0-12-187905-1. [Google Scholar]
- Rolfe, M.D.; Rice, C.J.; Lucchini, S.; Pin, C.; Thompson, A.; Cameron, A.D.S.; Alston, M.; Stringer, M.F.; Betts, R.P.; Baranyi, J.; et al. Lag Phase Is a Distinct Growth Phase That Prepares Bacteria for Exponential Growth and Involves Transient Metal Accumulation. J. Bacteriol. 2012, 194, 686–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Fan, D.; Chen, W.; Terentjev, E.M. Bacterial growth, detachment and cell size control on polyethylene terephthalate surfaces. Sci. Rep. 2015, 5, 15159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertrand, R.L. Lag Phase Is a Dynamic, Organized, Adaptive, and Evolvable Period That Prepares Bacteria for Cell Division. J. Bacteriol. 2019, 201, e00697-18. [Google Scholar] [CrossRef] [PubMed]
- Widdel, F. Theory and measurement of bacterial growth. Dalam Grund. Mikrobiol. 2007, 4, 1–11. [Google Scholar]
- Essendoubi, M.; Toubas, D.; Lepouse, C.; Leon, A.; Bourgeade, F.; Pinon, J.M.; Manfait, M.; Sockalingum, G.D. Epidemiological investigation and typing of Candida glabrata clinical isolates by FTIR spectroscopy. J. Microbiol. Methods 2007, 71, 325–331. [Google Scholar] [CrossRef]
- Maquelin, K.; Kirschner, C.; Choo-Smith, L.P.; Ngo-Thi, N.A.; van Vreeswijk, T.; Stämmler, M.; Endtz, H.P.; Bruining, H.A.; Naumann, D.; Puppels, G.J. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 2003, 41, 324–329. [Google Scholar] [CrossRef] [Green Version]
- Pebotuwa, S.; Kochan, K.; Peleg, A.; Wood, B.R.; Heraud, P. Influence of the Sample Preparation Method in Discriminating Candida spp. Using ATR-FTIR Spectroscopy. Molecules 2020, 25, 1551. [Google Scholar] [CrossRef] [Green Version]
- Potocki, L.; Depciuch, J.; Kuna, E.; Worek, M.; Lewinska, A.; Wnuk, M. FTIR and Raman Spectroscopy-Based Biochemical Profiling Reflects Genomic Diversity of Clinical Candida Isolates That May Be Useful for Diagnosis and Targeted Therapy of Candidiasis. Int. J. Mol. Sci. 2019, 20, 988. [Google Scholar] [CrossRef] [Green Version]
- De Bruyne, S.; Speeckaert, M.M.; Delanghe, J.R. Applications of mid-infrared spectroscopy in the clinical laboratory setting. Crit. Rev. Clin. Lab. Sci. 2018, 55, 1–20. [Google Scholar] [CrossRef]
- Kochan, K.; Nethercott, C.; Perez−Guaita, D.; Jiang, J.-H.; Peleg, A.Y.; Wood, B.R.; Heraud, P. Detection of Antimicrobial Resistance-Related Changes in Biochemical Composition of Staphylococcus aureus by Means of Atomic Force Microscopy-Infrared Spectroscopy. Anal. Chem. 2019, 91, 15397–15403. [Google Scholar] [CrossRef]
- Kochan, K.; Perez-Guaita, D.; Pissang, J.; Jiang, J.H.; Peleg, A.Y.; McNaughton, D.; Heraud, P.; Wood, B.R. In vivo atomic force microscopy-infrared spectroscopy of bacteria. J. R. Soc. Interface 2018, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novais, A.; Freitas, A.R.; Rodrigues, C.; Peixe, L. Fourier transform infrared spectroscopy: Unlocking fundamentals and prospects for bacterial strain typing. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 427–448. [Google Scholar] [CrossRef] [PubMed]
- Quintelas, C.; Ferreira, E.C.; Lopes, J.A.; Sousa, C. An Overview of the Evolution of Infrared Spectroscopy Applied to Bacterial Typing. Biotechnol. J. 2018, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitry, P.; Rebois, R.; Bourillot, E.; Deniset-Besseau, A.; Virolle, M.-J.; Lesniewska, E.; Dazzi, A. Combining infrared and mode synthesizing atomic force microscopy: Application to the study of lipid vesicles inside Streptomyces bacteria. Nano Res. 2016, 9, 1674–1681. [Google Scholar] [CrossRef]
- Pahlow, S.; Meisel, S.; Cialla-May, D.; Weber, K.; Rosch, P.; Popp, J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv. Drug Deliv. Rev. 2015, 89, 105–120. [Google Scholar] [CrossRef]
- Lorenz, B.; Wichmann, C.; Stockel, S.; Rosch, P.; Popp, J. Cultivation-Free Raman Spectroscopic Investigations of Bacteria. Trends Microbiol. 2017, 25, 413–424. [Google Scholar] [CrossRef]
- Zhao, X.; Li, M.; Xu, Z. Detection of Foodborne Pathogens by Surface Enhanced Raman Spectroscopy. Front. Microbiol. 2018, 9, 1236. [Google Scholar] [CrossRef]
- Mosier-Boss, P.A. Review on SERS of Bacteria. Biosensors 2017, 7, 51. [Google Scholar] [CrossRef] [Green Version]
- Oust, A.; Møretrø, T.; Kirschner, C.; Narvhus, J.A.; Kohler, A. Evaluation of the robustness of FT-IR spectra of lactobacilli towards changes in the bacterial growth conditions. FEMS Microbiol. Lett. 2004, 239, 111–116. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, R.; Verma, T.; Nandi, D.; Umapathy, S. Understanding the effects of culture conditions in bacterial growth: A biochemical perspective using Raman microscopy. J. Biophotonics 2020, 13, e201900233. [Google Scholar] [CrossRef]
- Al-qadiri, H.M.; Al-alami, N.I.; Lin, M.; Al-holy, M.; Cavinato, A.G.; Rasco, B.A. Studying of the bacterial growth phases using fourier transform infrared spectroscopy and multivariate analysis. J. Rapid Methods Autom. Microbiol. 2008, 16, 73–89. [Google Scholar] [CrossRef]
- Zeroual, W.; Choisy, C.; Doglia, S.M.; Bobichon, H.; Angiboust, J.-F.; Manfait, M. Monitoring of bacterial growth and structural analysis as probed by FT-IR spectroscopy. Biochim. Biophys. Acta Mol. Cell Res. 1994, 1222, 171–178. [Google Scholar] [CrossRef]
- Strola, S.; Marcoux, P.; Schultz, E.; Perenon, R.; Simon, A.-C.; Espagnon, I.; Allier, C.; Dinten, J. Differentiating the growth phases of single bacteria using Raman spectroscopy. In Proceedings of the Biomedical Vibrational Spectroscopy VI: Advances in Research and Industry, San Francisco, CA, USA, 4 March 2014; SPIE BiOS: Bellingham, WA, USA; Vol. 8939, p. 893905. [Google Scholar]
- Ren, Y.; Ji, Y.; Teng, L.; Zhang, H. Using Raman spectroscopy and chemometrics to identify the growth phase of Lactobacillus casei Zhang during batch culture at the single-cell level. Microb. Cell Fact. 2017, 16, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, C.; Bishop, M.M.; Barney, J.T.; Vargis, E. Effect of growth media and phase on Raman spectra and discrimination of mycobacteria. J. Biophotonics 2019, 12, e201900150. [Google Scholar] [CrossRef]
- Richardson, Z.; Perez-Guaita, D.; Kochan, K.; Wood, B.R. Determining the Age of Spoiled Milk from Dried Films Using Attenuated Reflection Fourier Transform Infrared (ATR FT-IR) Spectroscopy. Appl. Spectrosc. 2019, 73, 1041–1050. [Google Scholar] [CrossRef]
- Chatterjee, I.; Herrmann, M.; Proctor, R.A.; Peters, G.; Kahl, B.C. Enhanced Post-Stationary-Phase Survival of a Clinical Thymidine-Dependent Small-Colony Variant of Staphylococcus aureus Results from Lack of a Functional Tricarboxylic Acid Cycle. J. Bacteriol. 2007, 189, 2936–2940. [Google Scholar] [CrossRef] [Green Version]
- Missiakas, D.M.; Schneewind, O. Growth and laboratory maintenance of Staphylococcus aureus. Curr. Protoc. Microbiol. 2013, 8, 9C.1.1–9C.1.9. [Google Scholar] [CrossRef]

| IR Spectroscopy | Raman Spectroscopy | ||||
|---|---|---|---|---|---|
| Band Position [cm−1] | Assignment (Vibration) | Assignment (Compounds) | Band Position [cm−1] | Assignment (Vibration) | Assignment (Compounds) |
| 1745 | ν(C = O) | Lipids | 1666 | ν(C = C), Amide I | Lipids, Proteins |
| 1638 | Amide I | Proteins (β-sheet) | 1615 | ν(C = C), | Tyr, Trp |
| 1538 | Amide II | Proteins (β-sheet) | 1575 | Ring breathing (guanine, adenine) | Nucleic acids |
| 1515 | Tyrosine | 1480 | Ring breathing (guanine, adenine) | Nucleic acids | |
| 1452 | δas(CH3), δ(CH2), δ(CH)− | Proteins, Lipids, Polysaccharides | 1453 | δ(CH2), δ(CH3) | Protein |
| 1394 | δs(CH3) | Proteins | 1402 | δ(CH3) | Protein |
| 1335 | δ(CH), ring | Polysaccharides | 1384 | ||
| 1240 | νas(PO2)− Amide III | Nucleic acids, Phospholipids, Proteins | 1336 | Amide II, ring breathing (A,G) | Protein, Nucleic acids |
| 1215 | νas(PO2)− Amide III | Nucleic acids, Phospholipids, Proteins | 1244 | Amide III | Protein |
| 1117 | ν(C-C), ν(C-O), ν(P-O-C), | Polysaccharides, RNA | 1171 | δ(C-H) | Phe, Tyr |
| 1082 | νs(PO2)− | Nucleic acids, Phospholipids | 1098 | (PO2)− | Nucleic acids |
| 1057 | ν(C-O-C) | 1045 | ν(C-C), ν(C-C) | Polysaccharides | |
| 1032 | ν(C-C), ν(C-O), | Polysaccharides | 1005 | ν(C-C) | Phenylalanine |
| 994 | ν(C-C), ν(C-C) | Polysaccharides, ribose | 785 | O-P-O, ring breathing | Nucleic acids |
| 965 | ν(C-C), ν(C-C) | DNA | 729 | Ring breathing, A | Nucleic acids |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kochan, K.; Lai, E.; Richardson, Z.; Nethercott, C.; Peleg, A.Y.; Heraud, P.; Wood, B.R. Vibrational Spectroscopy as a Sensitive Probe for the Chemistry of Intra-Phase Bacterial Growth. Sensors 2020, 20, 3452. https://doi.org/10.3390/s20123452
Kochan K, Lai E, Richardson Z, Nethercott C, Peleg AY, Heraud P, Wood BR. Vibrational Spectroscopy as a Sensitive Probe for the Chemistry of Intra-Phase Bacterial Growth. Sensors. 2020; 20(12):3452. https://doi.org/10.3390/s20123452
Chicago/Turabian StyleKochan, Kamila, Elizabeth Lai, Zack Richardson, Cara Nethercott, Anton Y. Peleg, Philip Heraud, and Bayden R. Wood. 2020. "Vibrational Spectroscopy as a Sensitive Probe for the Chemistry of Intra-Phase Bacterial Growth" Sensors 20, no. 12: 3452. https://doi.org/10.3390/s20123452