Learning Diatoms Classification from a Dry Test Slide by Holographic Microscopy
Abstract
:1. Introduction
2. Materials and Methods
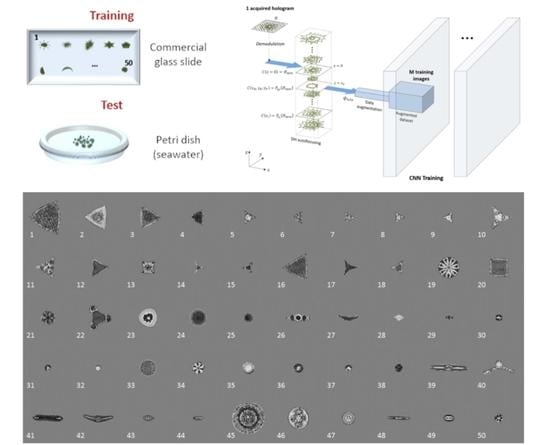
2.1. Holographic Acquisition: Training Based on a Commercial Glass Slide
2.2. Hologram Reconstruction and Data Augmentation
2.3. The Dataset
- 21 × 21 possible size scale in the range [−20%,+20%] × [−20%,+20%] around the initial QPI size by using the image resizing.
- 36 possible image orientation, by applying a 10 degree rotation step to the WQPI.
- 11 possible phase shift biases, taking in the uniformly distributed interval [0,π], that could be caused by random phase offsets during the recording and/or reconstruction processes (phase offsets as residual errors in the aberration compensation step).
2.4. Deep Learning Models
3. Experimental Results
4. Conclusions and Future Works
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shyamala, G.; Saravanakumar, N. Water quality assessment–a review. Int. J. Mod. Trends Eng. Sci. 2017, 4, 39–43. [Google Scholar]
- Wua, N.; Dong, X.; Liu, Y.; Wang, C.; Baattrup-Pedersen, A.; Riis, T. Using river microalgae as indicators for freshwater biomonitoring: Review of published research and future directions. Ecol. Indic. 2017, 81, 124–131. [Google Scholar] [CrossRef]
- Piper, J. A review of high-grade imaging of diatoms and radiolarians in light microscopy optical- and software-based techniques. Diatom Res. 2011, 26, 57–72. [Google Scholar] [CrossRef]
- Lopez, P.J.; Descles, J.; Allen, A.E.; Bowler, C. Prospects in diatom research. Curr. Opin. Biotechnol. 2005, 16, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Bedoshvili, Y.D.; Popkova, T.P.; Likhoshway, Y.V. Chloroplast structure of diatoms of different classes. Cell Tiss. Biol. 2009, 3, 297–310. [Google Scholar] [CrossRef]
- Du Buf, H. Diatom identification: A double challenge called ADIAC. In Proceedings of the 10th International Conference on Image Analysis and Processing, Venice, Italy, 31 October 1999; pp. 734–739. [Google Scholar]
- Du Buf, H.; Bayer, M. Automatic Diatom Identification. In Series in Machine Perception and Artificial Intelligence; World Scientific Publishing Co.: Munich, Germany, 2002. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 25; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; pp. 1097–1105. [Google Scholar]
- Jin, K.H.; McCann, M.T.; Froustey, E.; Unser, M. Deep Convolutional Neural Network for Inverse Problems in Imaging. IEEE Trans. Image Process. 2017, 26, 4509–4522. [Google Scholar] [CrossRef] [Green Version]
- Strack, R. Deep learning in imaging. Nat. Methods 2019, 16, 17. [Google Scholar] [CrossRef]
- Xing, F.; Xie, Y.; Su, H.; Liu, F.; Yang, L. Deep Learning in Microscopy Image Analysis: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 4550–4568. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.; Cho, H.; Lee, Y.S.; Choi, G.; Kim, G.; Min, H.; Park, Y. Quantitative Phase Imaging and Artificial Intelligence: A Review. IEEE J. Sel. Top. Quantum Electron. 2019, 25, 6800914. [Google Scholar] [CrossRef] [Green Version]
- Moen, E.; Bannon, D.; Kudo, T.; Graf, W.; Covert, M.; Van Valen, D. Deep learning for cellular image analysis. Nat. Methods 2019, 16, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Rivenson, Y.; Jin, Y.; Wei, Z.; Gao, R.; Günaydın, H.; Bentolila, L.A.; Kural, C.; Ozcan, A. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 2019, 16, 103–110. [Google Scholar] [CrossRef]
- Chen, C.L.; Mahjoubfar, A.; Tai, L.-C.; Blaby, I.K.; Huang, A.; Niazi, K.R.; Jalali, B. Deep Learning in Label-free Cell Classification. Sci. Rep. 2016, 6, 21471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miccio, L.; Cimmino, F.; Kurelac, I.; Villone, M.M.; Bianco, V.; Memmolo, P.; Merola, F.; Mugnano, M.; Capasso, M.; Iolascon, A.; et al. Perspectives on liquid biopsy for label-free detection of “circulating tumor cells” through intelligent lab-on-chips. View 2020, 1, 20200034. [Google Scholar] [CrossRef]
- Wang, Q.; Bi, S.; Sun, M.; Wang, Y.; Wang, D.; Yang, S. Deep learning approach to peripheral leukocyte recognition. PLoS ONE 2019, 14, e0218808. [Google Scholar] [CrossRef]
- Zeune, L.L.; Boink, Y.E.; Dalum, G.; Nanou, A.; De Wit, S.; Andree, K.C.; Swennenhuis, J.F.; Van Gils, S.A.; Terstappen, L.W.M.M.; Brune, C. Deep learning of circulating tumour cells. Nat. Mach. Intell. 2020, 2, 124–133. [Google Scholar] [CrossRef]
- Xu, M.; Papageorgiou, D.P.; Abidi, S.Z.; Dao, M.; Zhao, H.; Karniadakis, G.E. A deep convolutional neural network for classification of red blood cells in sickle cell anemia. PLoS Comput. Biol. 2017, 13, e1005746. [Google Scholar] [CrossRef]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-van de Kaa, C.; Bult, P.; Van Ginneken, B.; Van der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef] [Green Version]
- Pappas, J.; Stoermer, E. Legendre shape descriptors and shape group determination of specimens in the Cymbella cistula species complex. Phycologia 2003, 42, 90–97. [Google Scholar] [CrossRef] [Green Version]
- Dimitrovski, I.; Kocev, D.; Loskovska, S.; Džeroski, S. Hierarchical classification of diatom images using ensembles of predictive clustering trees. Ecol. Inform. 2012, 7, 19–29. [Google Scholar] [CrossRef]
- Bueno, G.; Deniz, O.; Pedraza, A.; Ruiz-Santaquiteria, J.; Salido, J.; Cristóbal, G.; Borrego-Ramos, M.; Blanco, S. Automated diatom classification (Part A): Handcrafted feature approaches. Appl. Sci. 2017, 7, 753. [Google Scholar] [CrossRef] [Green Version]
- Lai, Q.T.K.; Lee, K.C.M.; Tang, A.H.L.; Wong, K.K.Y.; So, H.K.H.; Tsia, K.K. High-throughput time-stretch imaging flow cytometry for multi-class classification of phytoplankton. Opt. Express 2016, 24, 28170–28184. [Google Scholar] [CrossRef]
- Pedraza, A.; Bueno, G.; Deniz, O.; Cristóbal, G.; Blanco, S.; Borrego-Ramos, M. Automated diatom classification (Part B): A deep learning approach. Appl. Sci. 2017, 7, 460. [Google Scholar] [CrossRef] [Green Version]
- Dunker, S.; Boho, D.; Wäldchen, J.; Mäder, P. Combining high-throughput imaging flow cytometry and deep learning for efficient species and life-cycle stage identification of phytoplankton. BMC Ecology 2018, 18, 51. [Google Scholar] [CrossRef] [Green Version]
- Zetsche, E.M.; El Mallahi, A.; Meysman, F.J.R. Digital holographic microscopy: A novel tool to study the morphology, physiology and ecology of diatoms. Diatom Res. 2016, 31, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Merola, F.; Memmolo, P.; Miccio, L.; Savoia, R.; Mugnano, M.; Fontana, A.; D’Ippolito, G.; Sardo, A.; Iolascon, A.; Gambale, A.; et al. Tomographic flow cytometry by digital holography. Light Sci. Appl. 2017, 6, e16241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umemura, K.; Matsukawa, Y.; Ide, Y.; Mayama, S. Label-free imaging and analysis of subcellular parts of a living diatom cylindrotheca sp. using optical diffraction tomography. MethodsX 2020, 7, 100889. [Google Scholar] [CrossRef] [PubMed]
- Merola, F.; Memmolo, P.; Miccio, L.; Bianco, V.; Paturzo, M.; Ferraro, P. Diagnostic tools for lab-on-chip applications based on coherent imaging microscopy. Proc. IEEE 2015, 103, 192–204. [Google Scholar] [CrossRef]
- Bianco, V.; Memmolo, P.; Carcagnì, P.; Merola, F.; Paturzo, M.; Distante, C.; Ferraro, P. Microplastic Identification via Holographic Imaging and Machine Learning. Adv. Intell. Syst. 2020, 2, 1900153. [Google Scholar] [CrossRef] [Green Version]
- Merola, F.; Memmolo, P.; Bianco, V.; Paturzo, M.; Mazzocchi, M.G.; Ferraro, P. Searching and identifying microplastics in marine environment by digital holography. Eur. Phys. J. Plus 2018, 133, 350. [Google Scholar] [CrossRef]
- Kloster, M.; Langenkämper, D.; Zurowietz, M.; Beszteri, B.; Nattkemper, T.W. Deep learning-based diatom taxonomy on virtual slides. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Cacace, T.; Bianco, V.; Mandracchia, B.; Pagliarulo, V.; Oleandro, E.; Paturzo, M.; Ferraro, P. Compact off-axis holographic slide microscope: Design guidelines. Biomed. Opt. Express 2020, 11, 2511–2532. [Google Scholar] [CrossRef] [PubMed]
- Talapatra, S.; Hong, J.; McFarland, M.; Nayak, A.R. Characterization of biophysical interactions in the water column using in situ digital holography. Mar. Ecol. Progress Ser. 2013, 473, 29–51. [Google Scholar] [CrossRef] [Green Version]
- Göröcs, Z.; Tamamitsu, M.; Bianco, V.; Wolf, P.; Roy, S.; Shindo, K.; Yanny, K.; Wu, Y.; Koydemir, H.C.; Rivenson, Y.; et al. A deep learning-enabled portable imaging flow cytometer for cost-effective, high-throughput, and label-free analysis of natural water samples. Light Sci. Appl. 2018, 7, 1–12. [Google Scholar] [CrossRef]
- Bianco, V.; Mandracchia, B.; Marchesano, V.; Pagliarulo, V.; Olivieri, F.; Coppola, S.; Ferraro, P. Endowing a plain fluidic chip with micro-optics: A holographic microscope slide. Light Sci. Appl. 2017, 6, e17055. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the Computer Vision and Pattern Recognition CVPR, Honolulu, HI, USA, 21–26 July 2017; Volume 1, p. 3. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Tan, M.; Le, Q.V. Efficientnet: Rethinking model scaling for convolutional neural networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- Radosavovic, I.; Kosaraju, R.P.; Girshick, R.; He, K.; Dollar, P. Designing network design spaces. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; Volume 10, pp. 428–436. [Google Scholar]
- Xie, S.; Girshick, R.; Doll’ar, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- Xu, W.; Jericho, M.H.; Meinertzhagen, I.A.; Kreuzer, H.J. Digital in-line holography for biological applications. Proc. Natl. Acad. Sci. USA 2001, 98, 11301–11305. [Google Scholar] [CrossRef] [Green Version]
- Watson, J.; Alexander, S.; Craig, G.; Hendry, D.C.; Hobson, P.R.; Lampitt, R.S.; Marteau, J.M.; Nareid, H.; Player, M.A.; Saw, K.; et al. Simultaneous in-line and off-axis subsea holographic recording of plankton and other marine particles. Meas. Sci. Tech. 2001, 12, L9. [Google Scholar] [CrossRef]
- 4-Deep. Holographic and Fluorescence microscopes. Available online: http://4-deep.com/ (accessed on 1 November 2020).
- Dyomin, V.; Davydova, A.; Morgalev, S.; Kirillov, N.; Olshukov, A.; Polovtsev, I.; Davydov, S. Monitoring of Plankton Spatial and Temporal Characteristics With the Use of a Submersible Digital Holographic Camera. Front. Mar. Sci. 2020, 28, 1–9. [Google Scholar]






| Model | Accuracy | Computational Time (Minutes) |
|---|---|---|
| EfficientNET-B0 | 0.91 | 414 |
| EfficientNET-B1 | 0.94 | 552 |
| EfficientNET-B2 | 0.88 | 588 |
| EfficientNET-B3 | 0.89 | 678 |
| EfficientNET-B7 | 0.72 | 3198 |
| ResNET50 | 0.89 | 455 |
| ResNET101 | 0.83 | 664 |
| SE-ResNET50 | 0.95 | 433 |
| SE-ResNET101 | 0.88 | 744 |
| SeNET154 | 0.83 | 5401 |
| DenseNET121 | 0.73 | 497 |
| RegNETY6.4GF | 0.85 | 1226 |
| RegNETY4.0GF | 0.80 | 650 |
| ENSEMBLE | 0.98 | --- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Memmolo, P.; Carcagnì, P.; Bianco, V.; Merola, F.; Goncalves da Silva Junior, A.; Garcia Goncalves, L.M.; Ferraro, P.; Distante, C. Learning Diatoms Classification from a Dry Test Slide by Holographic Microscopy. Sensors 2020, 20, 6353. https://doi.org/10.3390/s20216353
Memmolo P, Carcagnì P, Bianco V, Merola F, Goncalves da Silva Junior A, Garcia Goncalves LM, Ferraro P, Distante C. Learning Diatoms Classification from a Dry Test Slide by Holographic Microscopy. Sensors. 2020; 20(21):6353. https://doi.org/10.3390/s20216353
Chicago/Turabian StyleMemmolo, Pasquale, Pierluigi Carcagnì, Vittorio Bianco, Francesco Merola, Andouglas Goncalves da Silva Junior, Luis Marcos Garcia Goncalves, Pietro Ferraro, and Cosimo Distante. 2020. "Learning Diatoms Classification from a Dry Test Slide by Holographic Microscopy" Sensors 20, no. 21: 6353. https://doi.org/10.3390/s20216353