Intracellular Detection and Localization of Nanoparticles by Refractive Index Measurement
Abstract
:1. Introduction
2. Materials and Methods
2.1. Measurement of Refractive Indices
2.2. Cell Culture and Nanoparticle Preparation
2.3. Cell Toxicity Measurement
2.4. Statistical Analysis
3. Results
3.1. Nanodiamond Toxicity
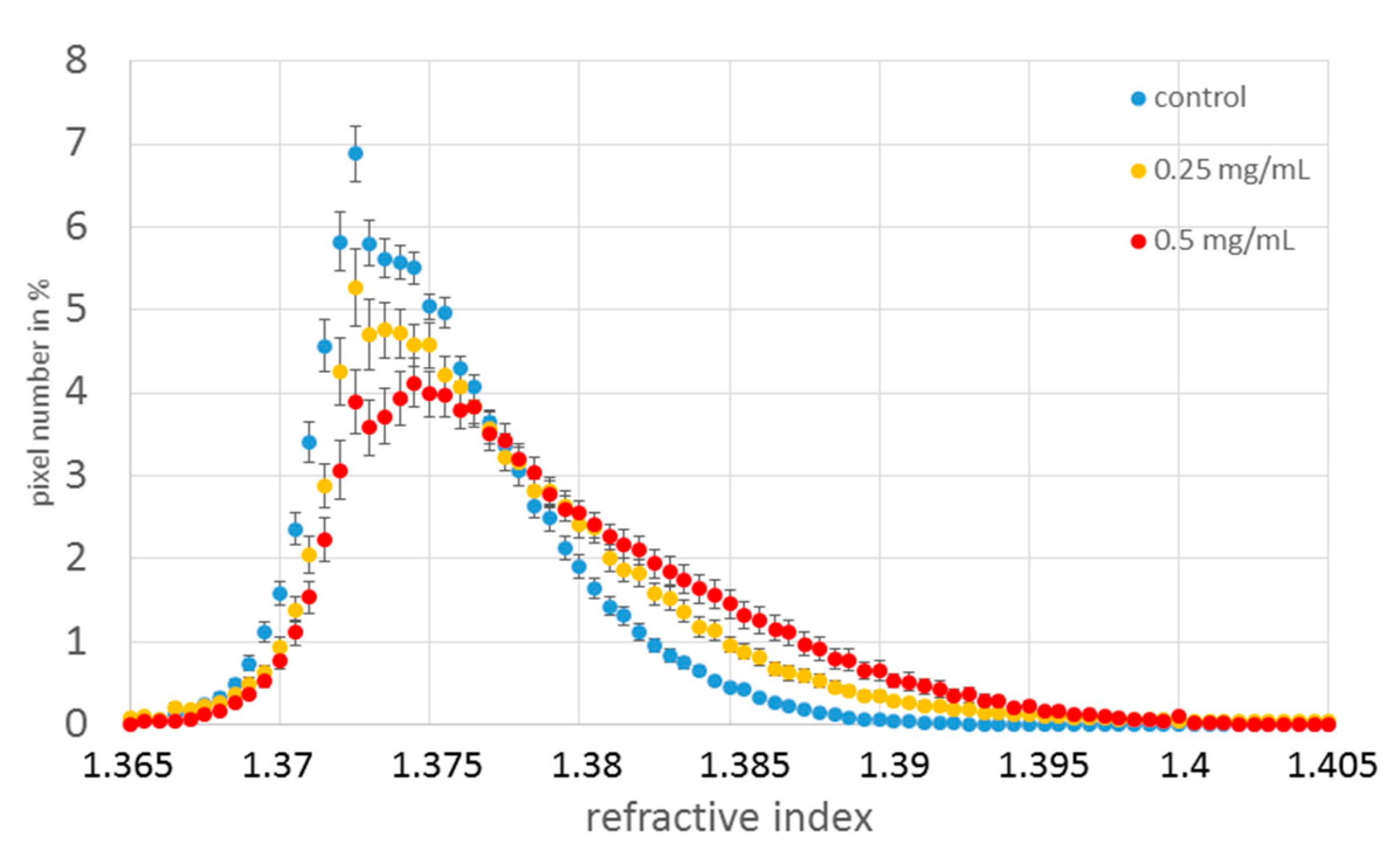
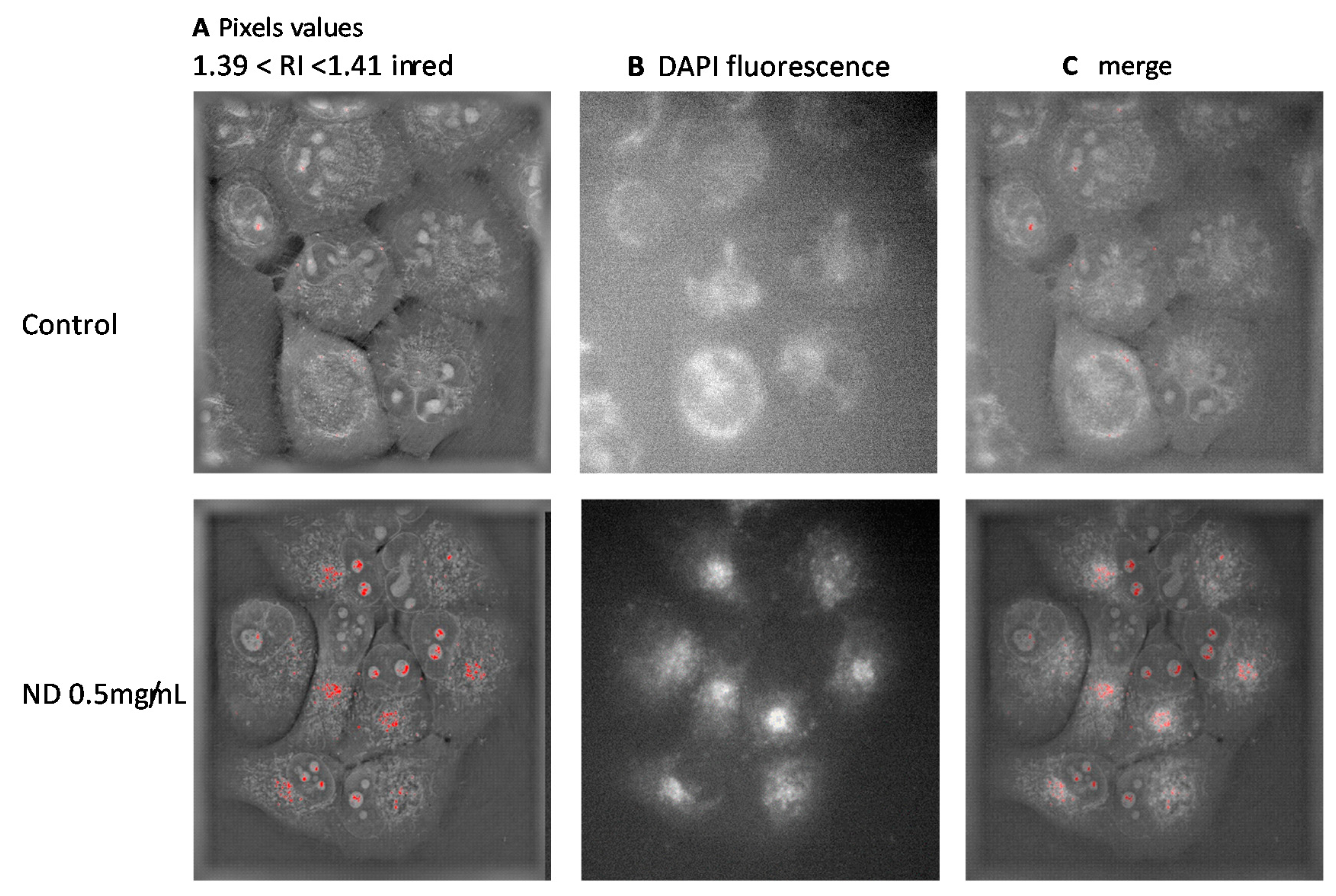
3.2. Refractive Index Measurement
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Jin, D.; Zhou, R.; Yaqoob, Z.; So, P.T.C. Tomographic Phase Microscopy: Principles and Applications in Bioimaging [Invited]. J. Opt. Soc. Am. B 2017, 34, B64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquet, P.; Rappaz, B.; Magistretti, P.J.; Cuche, E.; Emery, Y.; Colomb, T.; Depeursinge, C. Digital Holographic Microscopy: A Noninvasive Contrast Imaging Technique Allowing Quantitative Visualization of Living Cells with Subwavelength Axial Accuracy. Opt. Lett. 2005, 30, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquet, P.; Depeursinge, C.; Magistretti, P.J. Exploring Neural Cell Dynamics with Digital Holographic Microscopy. Annu. Rev. Biomed. Eng. 2013, 15, 407–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charrière, F.; Marian, A.; Montfort, F.; Kuehn, J.; Colomb, T.; Cuche, E.; Marquet, P.; Depeursinge, C. Cell Refractive Index Tomography by Digital Holographic Microscopy. Opt. Lett. 2006, 31, 178. [Google Scholar] [CrossRef] [PubMed]
- Cotte, Y.; Toy, F.; Jourdain, P.; Pavillon, N.; Boss, D.; Magistretti, P.; Marquet, P.; Depeursinge, C. Marker-Free Phase Nanoscopy. Nature Photon 2013, 7, 113–117. [Google Scholar] [CrossRef]
- Rappaz, B.; Marquet, P.; Cuche, E.; Emery, Y.; Depeursinge, C.; Magistretti, P.J. Measurement of the Integral Refractive Index and Dynamic Cell Morphometry of Living Cells with Digital Holographic Microscopy. Opt. Express 2005, 13, 9361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, X.J.; Liu, A.Q.; Lim, C.S.; Ayi, T.C.; Yap, P.H. Determining Refractive Index of Single Living Cell Using an Integrated Microchip. Sens. Actuators A Phys. 2007, 133, 349–354. [Google Scholar] [CrossRef]
- Choi, W.; Fang-Yen, C.; Badizadegan, K.; Oh, S.; Lue, N.; Dasari, R.R.; Feld, M.S. Tomographic Phase Microscopy. Nat. Methods 2007, 4, 717–719. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.Y.; Chin, L.K.; Ser, W.; Chen, H.F.; Hsieh, C.-M.; Lee, C.-H.; Sung, K.-B.; Ayi, T.C.; Yap, P.H.; Liedberg, B.; et al. Cell Refractive Index for Cell Biology and Disease Diagnosis: Past, Present and Future. Lab. Chip. 2016, 16, 634–644. [Google Scholar] [CrossRef] [PubMed]
- Tuchin, V.V. Tissue Optics: Light Scattering Methods and Instruments for Medical Diagnosis, 2nd ed.; Editor SPIE Press Book: Bellingham, WA, USA, 2007; Volume 13, 841p. [Google Scholar]
- Wang, P.; Bista, R.K.; Qiu, W.; Khalbuss, W.E.; Zhang, L.; Brand, R.E.; Liu, Y. An Insight into Statistical Refractive Index Properties of Cell Internal Structure via Low-Coherence Statistical Amplitude Microscopy. Opt. Express 2010, 18, 21950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bista, R.K.; Uttam, S.; Wang, P.; Staton, K.; Choi, S.; Bakkenist, C.J.; Hartman, D.J.; Brand, R.E.; Liu, Y. Quantification of Nanoscale Nuclear Refractive Index Changes during the Cell Cycle. J. Biomed. Opt. 2011, 16, 070503. [Google Scholar] [CrossRef]
- Chen, D.; Ganesh, S.; Wang, W.; Amiji, M. Plasma Protein Adsorption and Biological Identity of Systemically Administered Nanoparticles. Nanomedicine 2017, 12, 2113–2135. [Google Scholar] [CrossRef]
- Hinderliter, P.M.; Minard, K.R.; Orr, G.; Chrisler, W.B.; Thrall, B.D.; Pounds, J.G.; Teeguarden, J.G. ISDD: A Computational Model of Particle Sedimentation, Diffusion and Target Cell Dosimetry for in vitro Toxicity Studies. Part Fibre Toxicol. 2010, 7, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zousman, B.; Levinson, O. Pure nanodiamonds produced by laser-assisted technique, Chapter 5. In Nanodiamond Editor Oliver Williams; RSC Nanoscience & Nanotechnology: London, UK, 2014; pp. 112–127. [Google Scholar]
- Kim, D.; Oh, N.; Kim, K.; Lee, S.; Pack, C.-G.; Park, J.-H.; Park, Y. Label-Free High-Resolution 3-D Imaging of Gold Nanoparticles inside Live Cells Using Optical Diffraction Tomography. Methods 2018, 136, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Khanal, D.; Zhang, F.; Song, Y.; Hau, H.; Gautam, A.; Yamaguchi, S.; Uertz, J.; Mills, S.; Kondyurin, A.; Knowles, J.C.; et al. Biological Impact of Nanodiamond Particles—Label Free, High-Resolution Methods for Nanotoxicity Assessment. Nanotoxicology 2019, 13, 1210–1226. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Géloën, A.; Isaieva, K.; Isaiev, M.; Levinson, O.; Berger, E.; Lysenko, V. Intracellular Detection and Localization of Nanoparticles by Refractive Index Measurement. Sensors 2021, 21, 5001. https://doi.org/10.3390/s21155001
Géloën A, Isaieva K, Isaiev M, Levinson O, Berger E, Lysenko V. Intracellular Detection and Localization of Nanoparticles by Refractive Index Measurement. Sensors. 2021; 21(15):5001. https://doi.org/10.3390/s21155001
Chicago/Turabian StyleGéloën, Alain, Karyna Isaieva, Mykola Isaiev, Olga Levinson, Emmanuelle Berger, and Vladimir Lysenko. 2021. "Intracellular Detection and Localization of Nanoparticles by Refractive Index Measurement" Sensors 21, no. 15: 5001. https://doi.org/10.3390/s21155001





