Linear Combination Properties of the Phasor Space in Fluorescence Imaging
Abstract
:1. Introduction
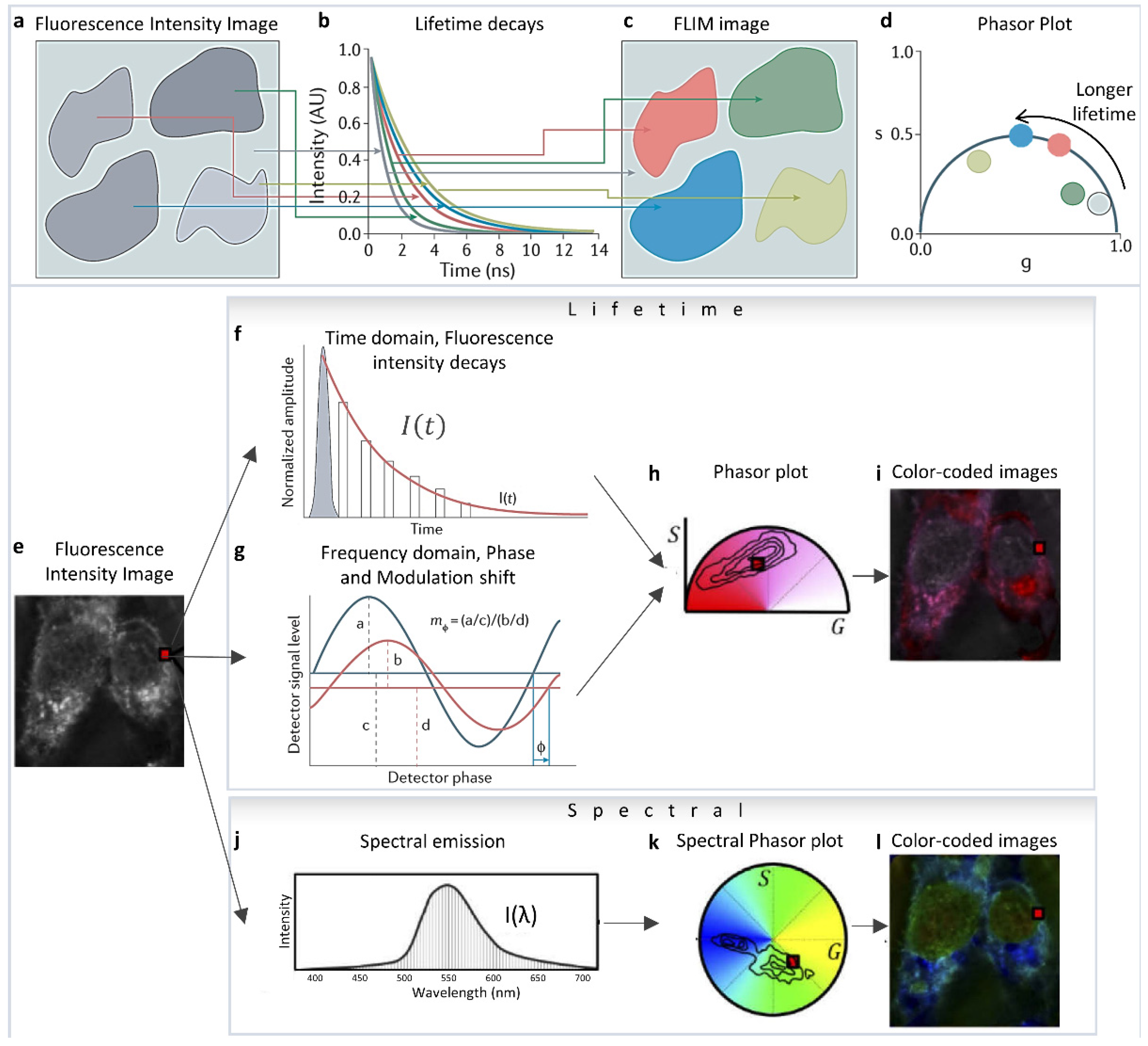
2. Mathematics
2.1. Mathematics of Phasor and Linear Combination in FLIM
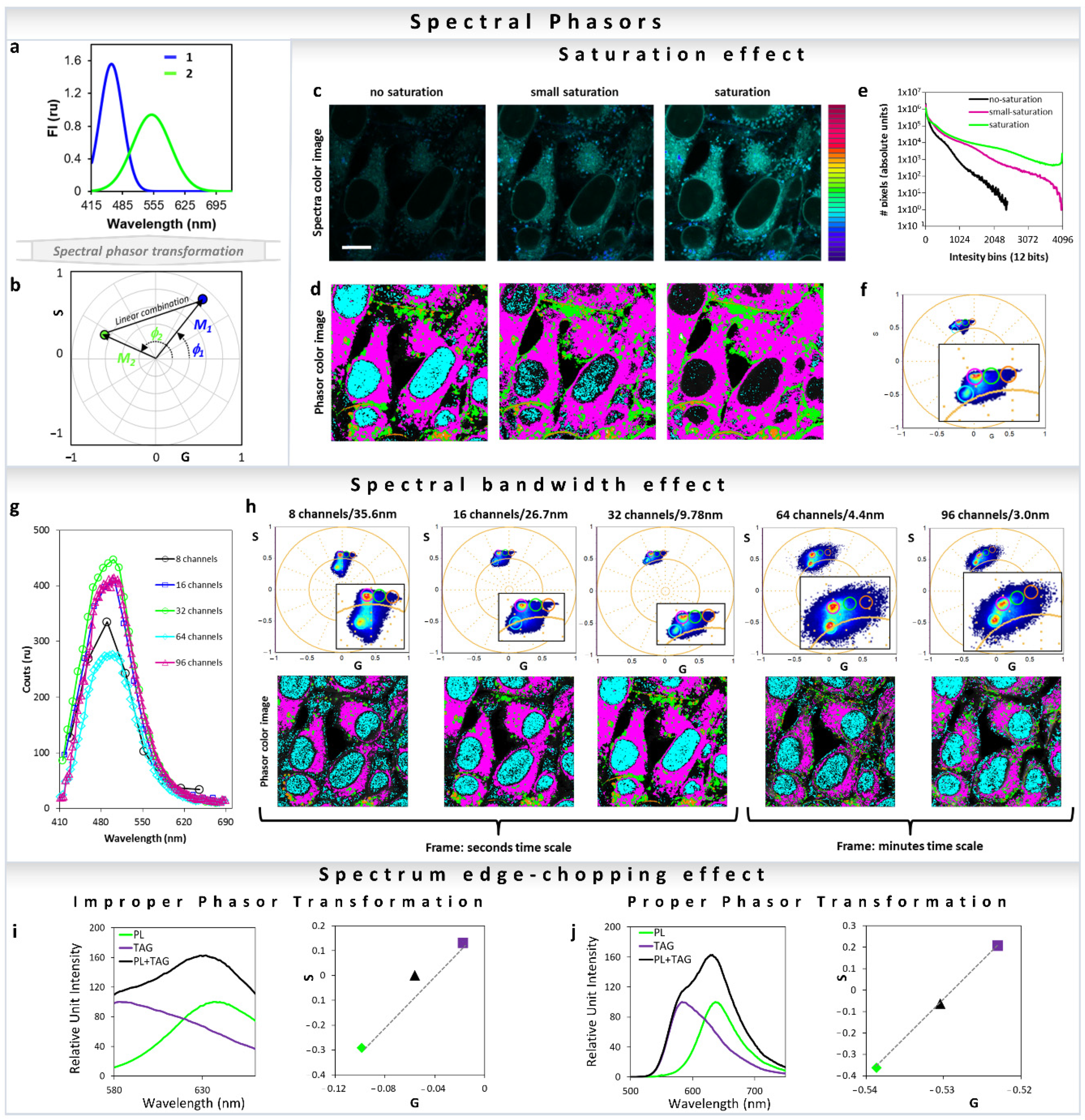
2.2. Mathematics of Spectral Phasor
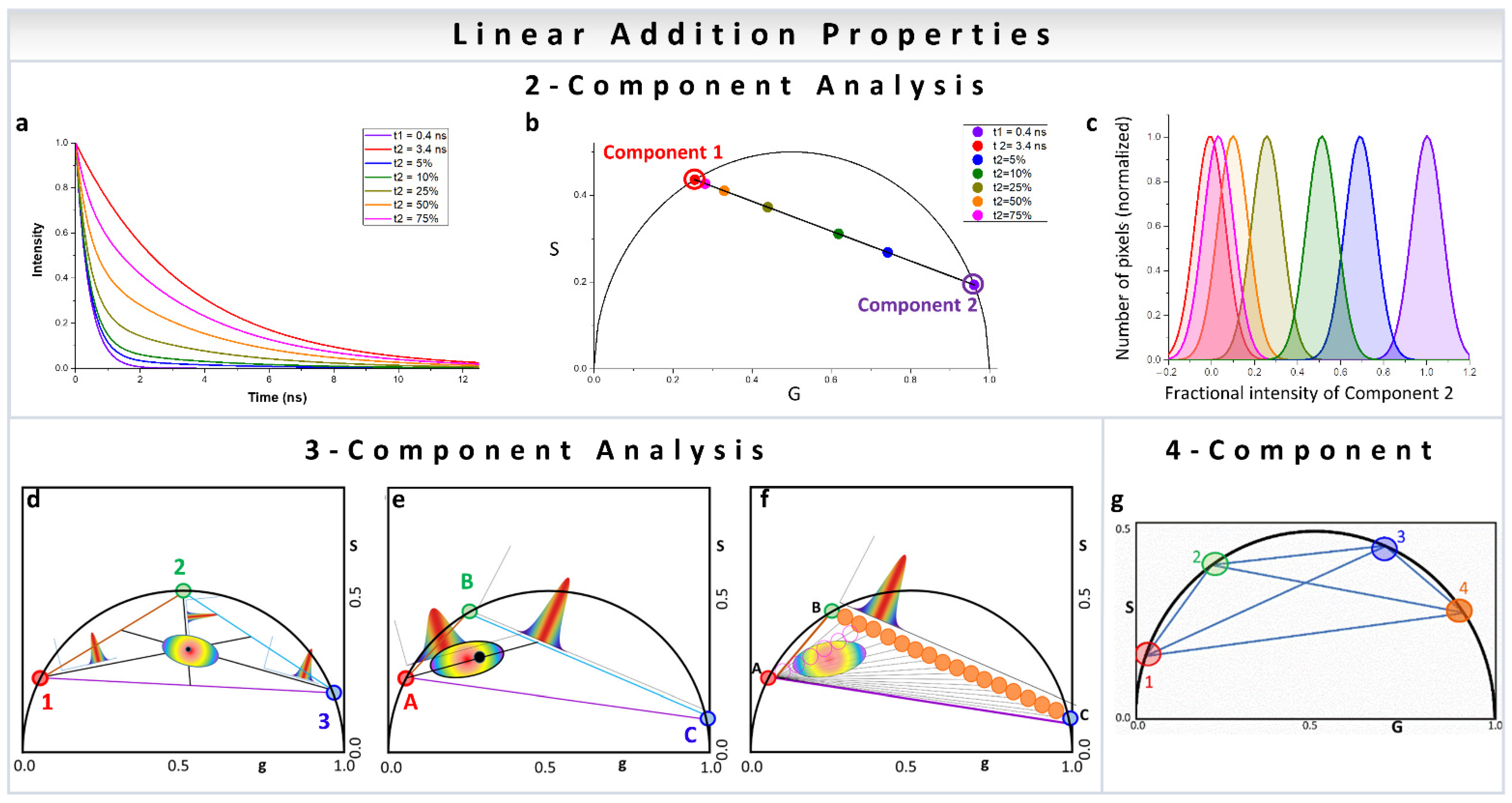
3. Component Analysis
3.1. Two-Component Phasor Analysis
3.2. Three-Component Analysis
3.3. Four-Component Measurement
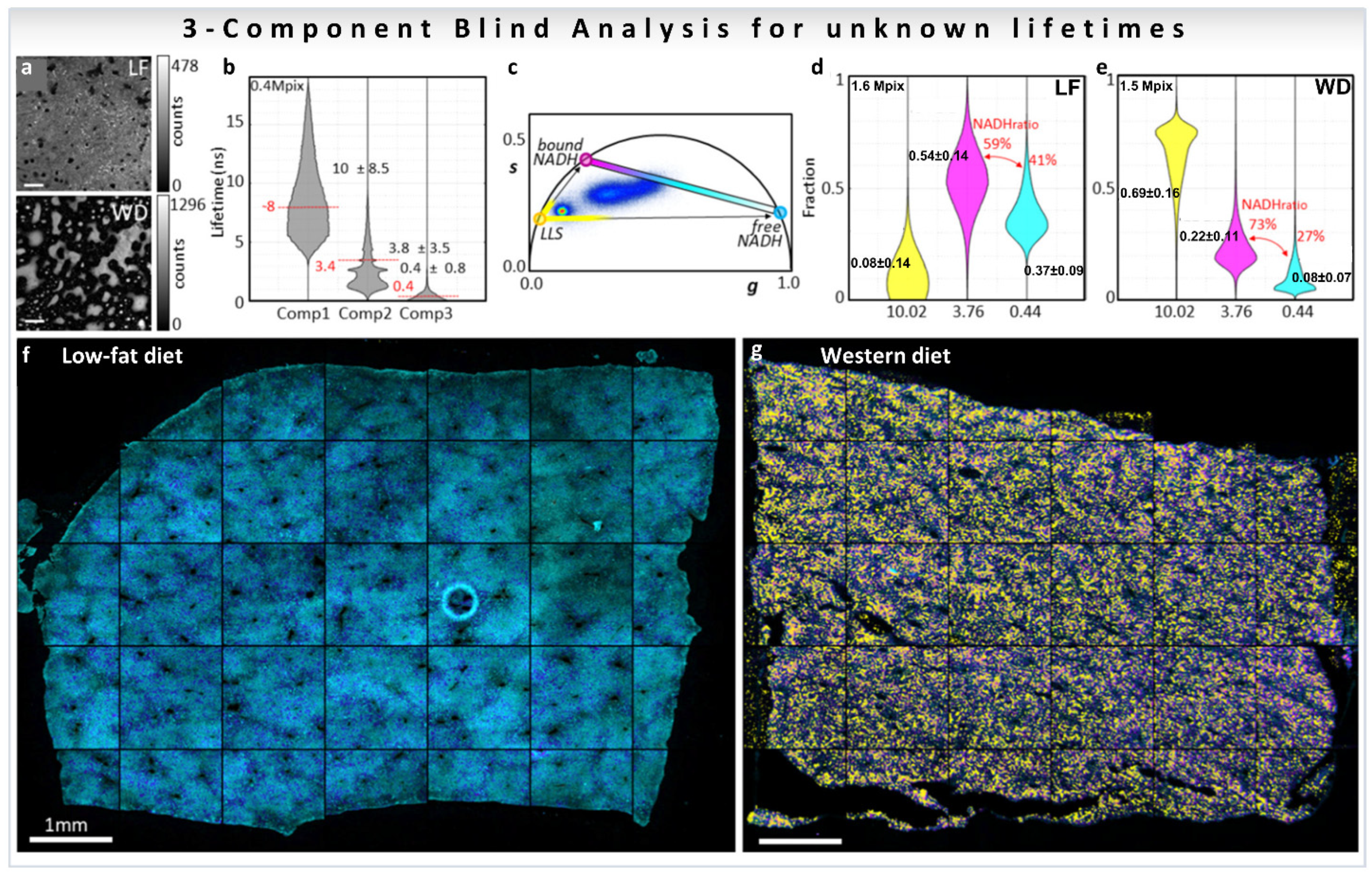
3.4. Blind Component Analysis
3.4.1. Two Components
3.4.2. Resolving N Components
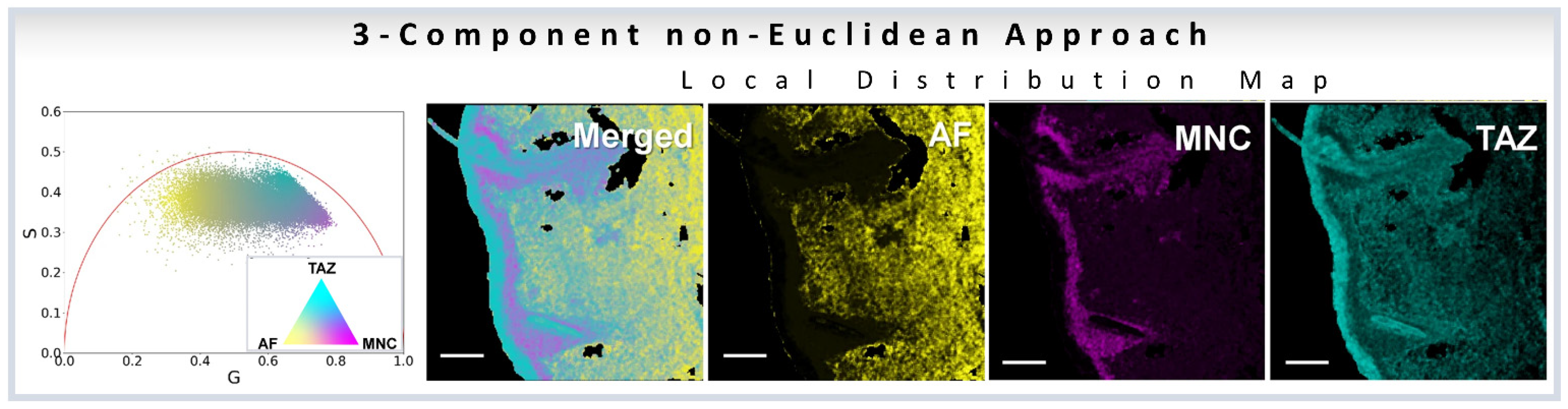
3.5. Three-Component Analysis Based on Non-Euclidean Approach
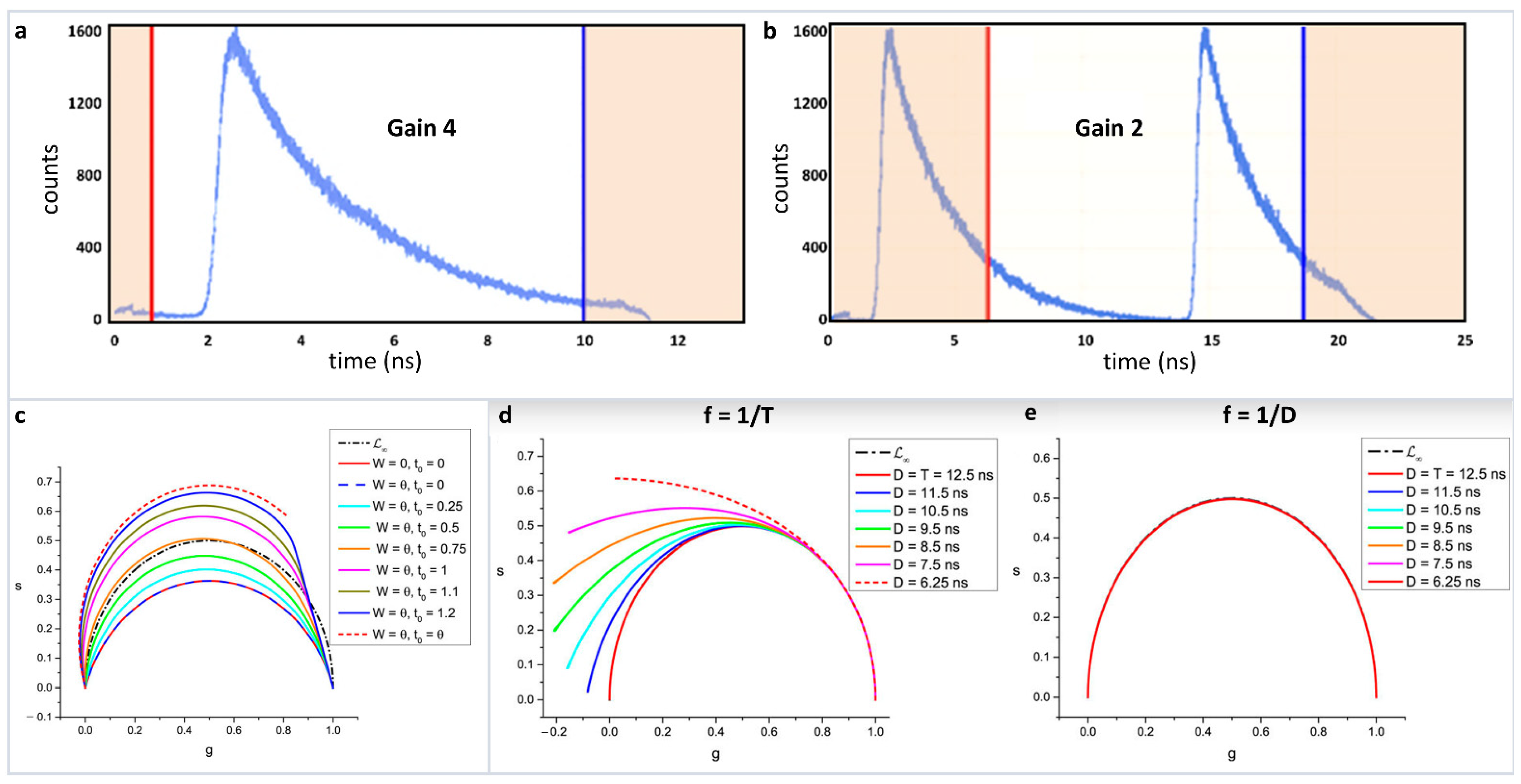
4. Breakdown of Linearity and Improper Transformation to Phasor Plot
5. Reciprocity Principle and Multidimensional Phasor Approach
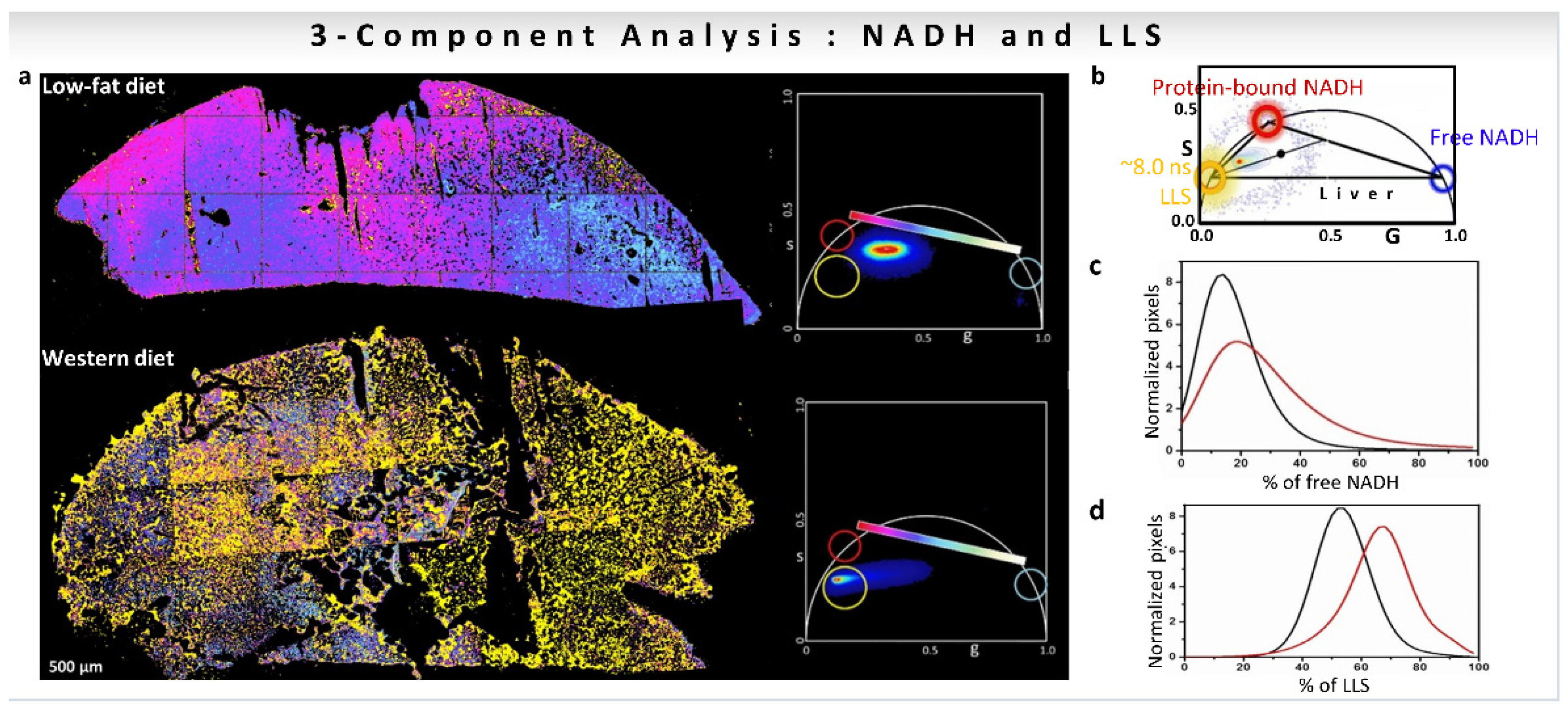
6. Results from Biological Measurements
6.1. Results of Multicomponent Analysis in Phasor-FLIM Imaging
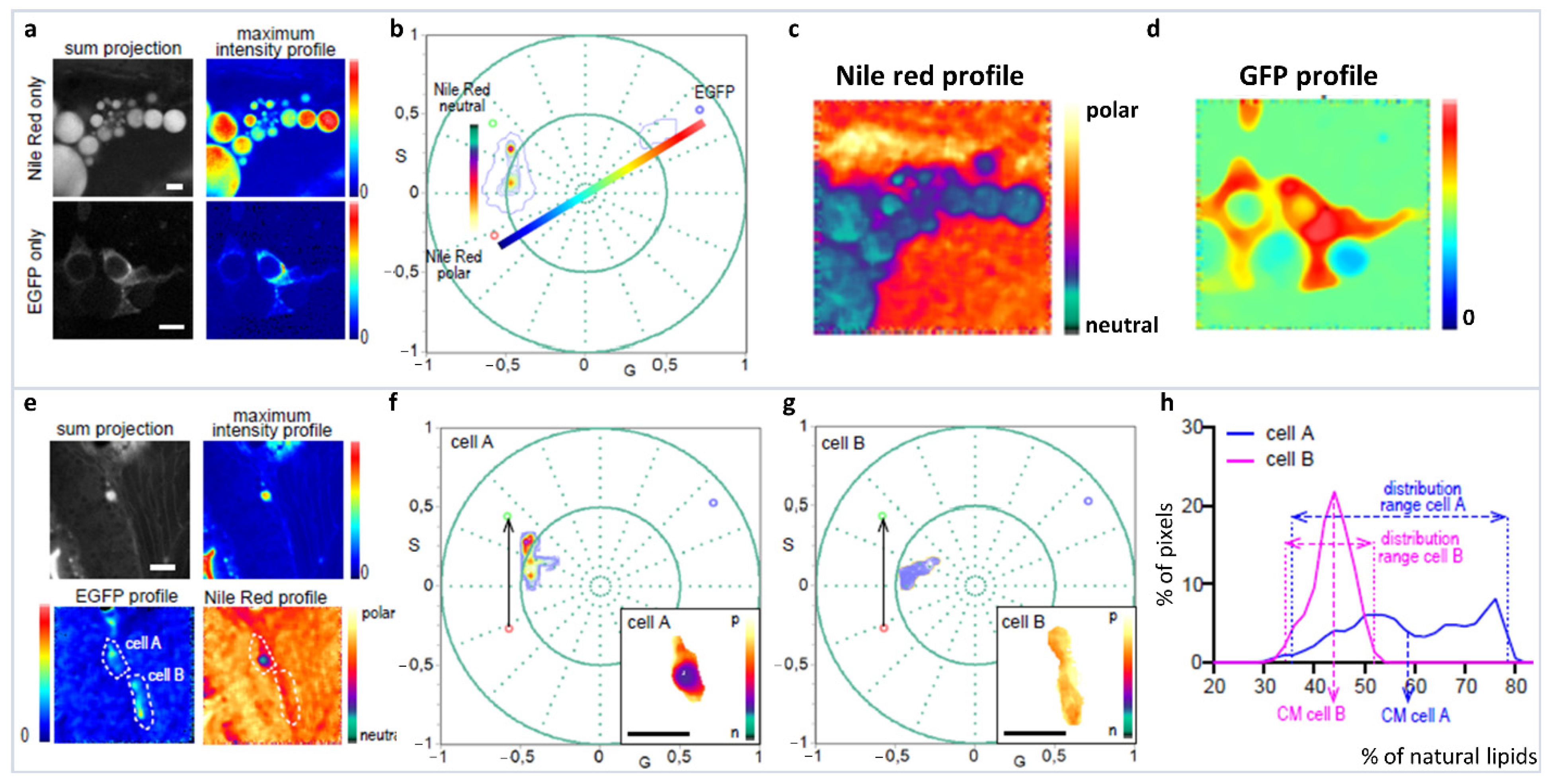
6.2. Spectral Phasor: Uses, Considerations and Applications
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malacrida, L.; Ranjit, S.; Jameson, D.M.; Gratton, E. The Phasor Plot: A Universal Circle To Advance Fluorescence Lifetime Analysis And Interpretation. Annu. Rev. Biophys. 2021, 50, 575–593. [Google Scholar] [CrossRef]
- Weber, G. Resolution of the Fluorescence Lifetimes in A Heterogeneous System by Phase and Modulation Measurements. J. Phys. Chem. 1981, 85, 949–953. [Google Scholar] [CrossRef]
- Jameson, D.M.; Gratton, E.; Hall, R.D. The Measurement and Analysis of Heterogeneous Emissions by Multifrequency Phase and Modulation Fluorometry. Appl Spectrosc. Rev. 1984, 20, 55–106. [Google Scholar] [CrossRef] [Green Version]
- Digman, M.A.; Caiolfa, V.R.; Zamai, M.; Gratton, E. The Phasor Approach to Fluorescence Lifetime Imaging Analysis. Biophys. J. 2008, 94, L14–L16. [Google Scholar] [CrossRef] [Green Version]
- Stringari, C.; Cinquin, A.; Cinquin, O.; Digman, M.A.; Donovan, P.J.; Gratton, E. Phasor Approach to Fluorescence Lifetime Microscopy Distinguishes Different Metabolic States of Germ Cells in A Live Tissue. Proc. Natl. Acad. Sci. USA 2011, 108, 13582–13587. [Google Scholar] [CrossRef] [Green Version]
- Stringari, C.; Nourse, J.L.; Flanagan, L.A.; Gratton, E. Phasor Fluorescence Lifetime Microscopy of Free and Protein-Bound Nadh Reveals Neural Stem Cell Differentiation Potential. PLoS ONE 2012, 7, E48014. [Google Scholar] [CrossRef] [Green Version]
- Hinde, E.; Digman, M.A.; Hahn, K.M.; Gratton, E. Millisecond Spatiotemporal Dynamics of Fret Biosensors by The Pair Correlation Function and The Phasor Approach to Flim. Proc. Natl. Acad. Sci. USA 2013, 110, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Celli, A.; Sanchez, S.; Behne, M.; Hazlett, T.; Gratton, E.; Mauro, T. The Epidermal Ca(2+) Gradient: Measurement Using The Phasor Representation of Fluorescent Lifetime Imaging. Biophys. J. 2010, 98, 911–921. [Google Scholar] [CrossRef] [Green Version]
- Redford, G.I.; Clegg, R.M. Polar Plot Representation for Frequency-Domain Analysis of Fluorescence Lifetimes. J. Fluoresc. 2005, 15, 805–815. [Google Scholar] [CrossRef]
- Eichorst, J.P.; Teng, K.W.; Clegg, R.M. Polar Plot Representation of Time-Resolved Fluorescence. In Fluorescence Spectroscopy and Microscopy: Methods and Protocols; Engelborghs, Y., Visser, A.J.W.G., Eds.; Humana Press: Totowa, NJ, USA, 2014; pp. 97–112. [Google Scholar]
- Clayton, A.H.; Hanley, Q.S.; Verveer, P.J. Graphical Representation and Multicomponent Analysis of Single-Frequency Fluorescence Lifetime Imaging Microscopy Data. J. Microsc. 2004, 213, 1–5. [Google Scholar] [CrossRef]
- Cervantes, M.; Forné, I.; Ranjit, S.; Gratton, E.; Imhof, A.; Sassone-Corsi, P. Bmal1 Associates with Nop58 in the Nucleolus and Contributes to Pre-Rrna Processing. Iscience 2020, 23, 101151. [Google Scholar] [CrossRef]
- Giral, H.; Cranston, D.; Lanzano, L.; Caldas, Y.; Sutherland, E.; Rachelson, J.; Dobrinskikh, E.; Weinman, E.J.; Doctor, R.B.; Gratton, E.; et al. Nhe3 Regulatory Factor 1 (Nherf1) Modulates Intestinal Sodium-Dependent Phosphate Transporter (Napi-2b) Expression in Apical Microvilli. J. Biol. Chem. 2012, 287, 35047–35056. [Google Scholar] [CrossRef] [Green Version]
- Giral, H.; Lanzano, L.; Caldas, Y.; Blaine, J.; Verlander, J.W.; Lei, T.; Gratton, E.; Levi, M. Role of Pdzk1 Protein in Apical Membrane Expression of Renal Sodium-Coupled Phosphate Transporters. J. Biol. Chem. 2011, 286, 15032–15042. [Google Scholar] [CrossRef] [Green Version]
- Greco, C.M.; Cervantes, M.; Fustin, J.-M.; Ito, K.; Ceglia, N.; Samad, M.; Shi, J.; Koronowski, K.B.; Forne, I.; Ranjit, S.; et al. S-Adenosyl-L-Homocysteine Hydrolase Links Methionine Metabolism to the Circadian Clock and Chromatin Remodeling. Sci. Adv. 2020, 6, Eabc5629. [Google Scholar] [CrossRef]
- Hinde, E.; Digman, M.A.; Welch, C.; Hahn, K.M.; Gratton, E. Biosensor Förster Resonance Energy Transfer Detection by the Phasor Approach to Fluorescence Lifetime Imaging Microscopy. Microsc. Res. Tech. 2012, 75, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Arnal, L.; Ranjit, S.; Stringari, C.; Orozco-Solis, R.; Gratton, E.; Sassone-Corsi, P. Spatial Dynamics of Sirt1 and the Subnuclear Distribution of Nadh Species. Proc. Natl. Acad. Sci. USA 2016, 113, 12715–12720. [Google Scholar] [CrossRef] [Green Version]
- Alfonso-García, A.; Smith, T.D.; Datta, R.; Luu, T.U.; Gratton, E.; Potma, E.O.; Liu, W.F. Label-Free Identification of Macrophage Phenotype by Fluorescence Lifetime Imaging Microscopy. J. Biomed. Opt. 2016, 21, 46005. [Google Scholar] [CrossRef]
- Datta, R.; Alfonso-García, A.; Cinco, R.; Gratton, E. Fluorescence Lifetime Imaging of Endogenous Biomarker of Oxidative Stress. Sci. Rep. 2015, 5, 9848. [Google Scholar] [CrossRef] [Green Version]
- Datta, R.; Heylman, C.; George, S.C.; Gratton, E. Label-Free Imaging of Metabolism and Oxidative Stress in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Biomed. Opt. Express 2016, 7, 1690–1701. [Google Scholar] [CrossRef] [Green Version]
- Sobrino, A.; Phan, D.T.; Datta, R.; Wang, X.; Hachey, S.J.; Romero-López, M.; Gratton, E.; Lee, A.P.; George, S.C.; Hughes, C.C. 3d Microtumors In Vitro Supported by Perfused Vascular Networks. Sci. Rep. 2016, 6, 31589. [Google Scholar] [CrossRef] [Green Version]
- Stringari, C.; Edwards, R.A.; Pate, K.T.; Waterman, M.L.; Donovan, P.J.; Gratton, E. Metabolic Trajectory of Cellular Differentiation in Small Intestine by Phasor Fluorescence Lifetime Microscopy of Nadh. Sci. Rep. 2012, 2, 568. [Google Scholar] [CrossRef] [Green Version]
- Stringari, C.; Sierra, R.; Donovan, P.J.; Gratton, E. Label-Free Separation of Human Embryonic Stem Cells and Their Differentiating Progenies by Phasor Fluorescence Lifetime Microscopy. J. Biomed. Opt. 2012, 17, 046012. [Google Scholar] [CrossRef] [Green Version]
- Trinh, A.L.; Chen, H.; Chen, Y.; Hu, Y.; Li, Z.; Siegel, E.R.; Linskey, M.E.; Wang, P.H.; Digman, M.A.; Zhou, Y.H. Tracking Functional Tumor Cell Subpopulations of Malignant Glioma by Phasor Fluorescence Lifetime Imaging Microscopy of Nadh. Cancers 2017, 9, 168. [Google Scholar] [CrossRef] [Green Version]
- Fereidouni, F.; Bader, A.N.; Gerritsen, H.C. Spectral Phasor Analysis Allows Rapid and Reliable Unmixing of Fluorescence Microscopy Spectral Images. Opt. Express 2012, 20, 12729–12741. [Google Scholar] [CrossRef] [Green Version]
- Malacrida, L.; Gratton, E. Laurdan Fluorescence and Phasor Plots Reveal the Effects of A H2O2 Bolus in Nih-3t3 Fibroblast Membranes Dynamics and Hydration. Free Radic. Biol. Med. 2018, 128, 144–156. [Google Scholar] [CrossRef]
- Ranjit, S.; Lanzanò, L.; Libby, A.E.; Gratton, E.; Levi, M. Advances in Fluorescence Microscopy Techniques to Study Kidney Function. Nat. Rev. Nephrol. 2021, 17, 128–144. [Google Scholar] [CrossRef]
- Jameson, D.M. Introduction to Fluorescence; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Digman, M.; Gratton, E. The Phasor Approach to Fluorescence Lifetime Imaging: Exploiting Phasor Linear Properties. In Fluorescence Lifetime Spectroscopy and Imaging; CRC Press: Boca Raton, FL, USA, 2014; pp. 235–248. [Google Scholar]
- Vallmitjana, A.; Torrado, B.; Gratton, E. Phasor-Based Image Segmentation: Machine Learning Clustering Techniques. Biomed. Opt. Express 2021, 12, 3410–3422. [Google Scholar] [CrossRef]
- Ranjit, S.; Malacrida, L.; Jameson, D.M.; Gratton, E. Fit-Free Analysis of Fluorescence Lifetime Imaging Data Using the Phasor Approach. Nat. Protoc. 2018, 13, 1979–2004. [Google Scholar] [CrossRef]
- Ranjit, S.; Datta, R.; Dvornikov, A.; Gratton, E. Multicomponent Analysis of Phasor Plot in A Single Pixel to Calculate Changes of Metabolic Trajectory in Biological Systems. J. Phys. Chem. A 2019, 123, 9865–9873. [Google Scholar] [CrossRef]
- Ranjit, S.; Dvornikov, A.; Dobrinskikh, E.; Wang, X.; Luo, Y.; Levi, M.; Gratton, E. Measuring The Effect of A Western Diet on Liver Tissue Architecture by Flim Autofluorescence and Harmonic Generation Microscopy. Biomed. Opt. Express 2017, 8, 3143–3154. [Google Scholar] [CrossRef] [Green Version]
- Ranjit, S.; Malacrida, L.; Stakic, M.; Gratton, E. Determination of the Metabolic Index Using the Fluorescence Lifetime of Free and Bound Nicotinamide Adenine Dinucleotide Using the Phasor Approach. J. Biophotonics 2019, 12, E201900156. [Google Scholar] [CrossRef]
- Vallmitjana, A.; Dvornikov, A.; Torrado, B.; Jameson, D.M.; Ranjit, S.; Gratton, E. Resolution of 4 Components in the Same Pixel in Flim Images Using the Phasor Approach. Methods Appl. Fluoresc. 2020, 8, 035001. [Google Scholar] [CrossRef] [Green Version]
- Jeong, S.; Greenfield, D.A.; Hermsmeier, M.; Yamamoto, A.; Chen, X.; Chan, K.F.; Evans, C.L. Time-Resolved Fluorescence Microscopy with Phasor Analysis for Visualizing Multicomponent Topical Drug Distribution within Human Skin. Sci. Rep. 2020, 10, 5360. [Google Scholar] [CrossRef] [Green Version]
- Osseiran, S.; Roider, E.; Wang, H.; Suita, Y.; Murphy, M.; Fisher, D.; Evans, C. Non-Euclidean Phasor Analysis for Quantification of Oxidative Stress In Ex Vivo Human Skin Exposed to Sun Filters Using Fluorescence Lifetime Imaging Microscopy. J. Biomed. Opt. 2017, 22, 125004. [Google Scholar] [CrossRef] [Green Version]
- Vallmitjana, A.; Torrado, B.; Dvornikov, A.; Ranjit, S.; Gratton, E. Blind Resolution of Lifetime Components in Individual Pixels of Fluorescence Lifetime Images Using the Phasor Approach. J. Phys. Chem. B 2020, 124, 10126–10137. [Google Scholar] [CrossRef]
- Michalet, X. Continuous and Discrete Phasor Analysis of Binned or Time-Gated Periodic Decays. Aip. Adv. 2021, 11, 035331. [Google Scholar] [CrossRef]
- Ranjit, S.; Malacrida, L.; Gratton, E. Differences between Flim Phasor Analyses for Data Collected with The Becker and Hickl Spc830 Card and with the Flimbox Card. Microsc. Res. Tech. 2018, 81, 980–989. [Google Scholar] [CrossRef]
- Malacrida, L.; Jameson, D.M.; Gratton, E. A Multidimensional Phasor Approach Reveals Laurdan Photophysics in Nih-3t3 Cell Membranes. Sci. Rep. 2017, 7, 9215. [Google Scholar] [CrossRef]
- Malacrida, L.; Gratton, E.; Jameson, D.M. Model-Free. Methods to Study Membrane Environmental Probes: A Comparison of the Spectral Phasor and Generalized Polarization Approaches. Methods Appl. Fluoresc. 2015, 3, 047001. [Google Scholar] [CrossRef]
- Malacrida, L.; Astrada, S.; Briva, A.; Bollati-Fogolín, M.; Gratton, E.; Bagatolli, L.A. Spectral Phasor Analysis of Laurdan Fluorescence in Live A549 Lung Cells to Study the Hydration and Time Evolution of Intracellular Lamellar Body-Like Structures. Biochim. Biophys. Acta 2016, 1858, 2625–2635. [Google Scholar] [CrossRef]
- Sameni, S.; Malacrida, L.; Tan, Z.; Digman, M.A. Alteration in Fluidity of Cell Plasma Membrane in Huntington Disease Revealed by Spectral Phasor Analysis. Sci. Rep. 2018, 8, 734. [Google Scholar] [CrossRef] [Green Version]
- Sena, F.; Sotelo-Silveira, M.; Astrada, S.; Botella, M.A.; Malacrida, L.; Borsani, O. Spectral Phasor Analysis Reveals Altered Membrane Order and Function of Root Hair Cells in Arabidopsis Dry2/Sqe1-5 Drought Hypersensitive Mutant. Plant. Physiol. BioChem. 2017, 119, 224–231. [Google Scholar] [CrossRef]
- Ranjit, S.; Lanzano, L.; Gratton, E. Mapping Diffusion in A Living Cell Via the Phasor Approach. Biophys. J. 2014, 107, 2775–2785. [Google Scholar] [CrossRef] [Green Version]
- Scott, T.G.; Spencer, R.D.; Leonard, N.J.; Weber, G. Synthetic Spectroscopic Models Related to Coenzymes and Base Pairs. V. Emission Properties of Nadh. Studies of Fluorescence Lifetimes and Quantum Efficiencies of Nadh, Acpyadh, [Reduced Acetylpyridineadenine Dinucleotide] and Simplified Synthetic Models. J. Am. Chem. Soc. 1970, 92, 687–695. [Google Scholar] [CrossRef]
- Shen, B.; Yan, J.; Wang, S.; Zhou, F.; Zhao, Y.; Hu, R.; Qu, J.; Liu, L. Label-Free Whole-Colony Imaging and Metabolic Analysis of Metastatic Pancreatic Cancer by An Autoregulating Flexible Optical System. Theranostics 2020, 10, 1849–1860. [Google Scholar] [CrossRef]
- Bird, D.K.; Yan, L.; Vrotsos, K.M.; Eliceiri, K.W.; Vaughan, E.M.; Keely, P.J.; White, J.G.; Ramanujam, N. Metabolic Mapping of Mcf10a Human Breast Cells via Multiphoton Fluorescence Lifetime Imaging of the Coenzyme Nadh. Cancer Res. 2005, 65, 8766–8773. [Google Scholar] [CrossRef] [Green Version]
- Skala, M.C.; Riching, K.M.; Bird, D.K.; Gendron-Fitzpatrick, A.; Eickhoff, J.; Eliceiri, K.W.; Keely, P.J.; Ramanujam, N. In Vivo Multiphoton Fluorescence Lifetime Imaging of Protein-Bound and Free Nicotinamide Adenine Dinucleotide in Normal and Precancerous Epithelia. J. Biomed. Opt. 2007, 12, 024014. [Google Scholar] [CrossRef] [Green Version]
- Skala, M.C.; Riching, K.M.; Gendron-Fitzpatrick, A.; Eickhoff, J.; Eliceiri, K.W.; White, J.G.; Ramanujam, N. In Vivo Multiphoton Microscopy of Nadh and Fad Redox States, Fluorescence Lifetimes, and Cellular Morphology in Precancerous Epithelia. Proc. Natl. Acad. Sci. USA 2007, 104, 19494–19499. [Google Scholar] [CrossRef] [Green Version]
- Kolenc, O.I.; Quinn, K.P. Evaluating Cell Metabolism through Autofluorescence Imaging of Nad(P)H and Fad. Antioxid. Redox Signal. 2019, 30, 875–889. [Google Scholar] [CrossRef]
- Ma, N.; Digman, M.A.; Malacrida, L.; Gratton, E. Measurements of Absolute Concentrations of Nadh in Cells Using the Phasor Flim Method. Biomed. Opt. Express 2016, 7, 2441–2452. [Google Scholar] [CrossRef] [Green Version]
- Wright, B.K.; Andrews, L.M.; Markham, J.; Jones, M.R.; Stringari, C.; Digman, M.A.; Gratton, E. Nadh Distribution in Live Progenitor Stem Cells by Phasor-Fluorescence Lifetime Image Microscopy. Biophys. J. 2012, 103, L7–L9. [Google Scholar] [CrossRef] [Green Version]
- Ferri, G.; Tesi, M.; Massarelli, F.; Marselli, L.; Marchetti, P.; Cardarelli, F. Metabolic Response of Insulinoma 1e Cells to Glucose Stimulation Studied by Fluorescence Lifetime Imaging. FASEB Bioadv. 2020, 2, 409–418. [Google Scholar] [CrossRef]
- Lepanto, P.; Levin-Ferreyra, F.; Koziol, U.; Malacrida, L.; Badano, J.L. Insights into in vivo adipocyte differentiation through cell-specific labeling in zebrafish. Biology Open 2021, 10, bio058734. [Google Scholar] [CrossRef]
- Parasassi, T.; De Stasio, G.; Ravagnan, G.; Rusch, R.M.; Gratton, E. Quantitation of Lipid Phases in Phospholipid Vesicles by the Generalized Polarization of Laurdan Fluorescence. Biophys. J. 1991, 60, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Franssen, W.M.J.; Vergeldt, F.J.; Bader, A.N.; Van Amerongen, H.; Terenzi, C. Full-Harmonics Phasor Analysis: Unravelling Multiexponential Trends in Magnetic Resonance Imaging Data. J. Phys. Chem. Lett. 2020, 11, 9152–9158. [Google Scholar] [CrossRef]
- Fu, D.; Xie, X.S. Reliable Cell Segmentation Based on Spectral Phasor Analysis of Hyperspectral Stimulated Raman Scattering Imaging Data. Anal. Chem. 2014, 86, 4115–4119. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torrado, B.; Malacrida, L.; Ranjit, S. Linear Combination Properties of the Phasor Space in Fluorescence Imaging. Sensors 2022, 22, 999. https://doi.org/10.3390/s22030999
Torrado B, Malacrida L, Ranjit S. Linear Combination Properties of the Phasor Space in Fluorescence Imaging. Sensors. 2022; 22(3):999. https://doi.org/10.3390/s22030999
Chicago/Turabian StyleTorrado, Belén, Leonel Malacrida, and Suman Ranjit. 2022. "Linear Combination Properties of the Phasor Space in Fluorescence Imaging" Sensors 22, no. 3: 999. https://doi.org/10.3390/s22030999





