Wavelet-Based Fractal Analysis of rs-fMRI for Classification of Alzheimer’s Disease
Abstract
:1. Introduction
1.1. Alzheimer’s Disease Classification—Related Work
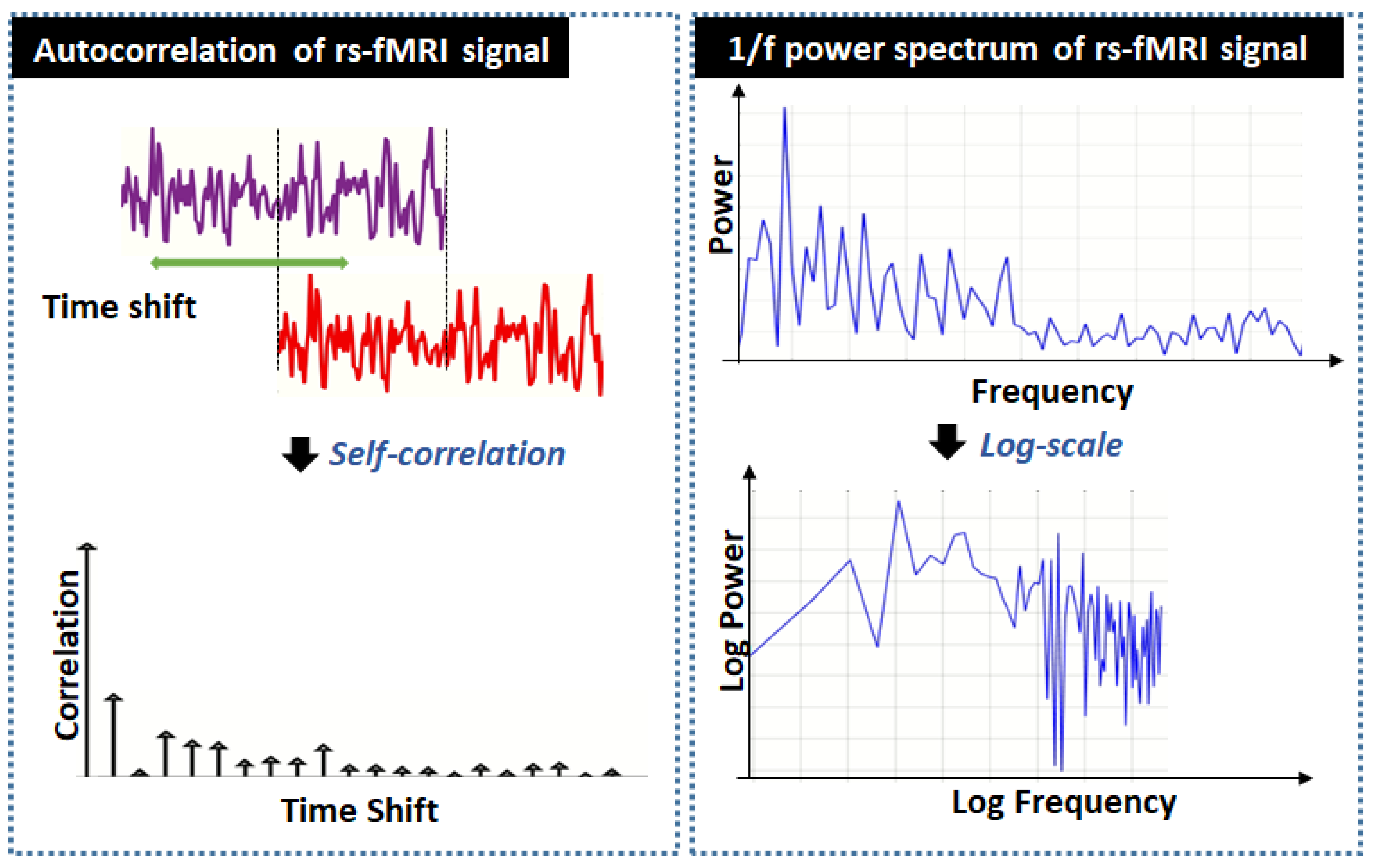
1.2. Fractal Behavior of rs-fMRI Signals
2. Long Memory Model of rs-fMRI Signals
2.1. Univariate Case
2.2. Multivariate Case
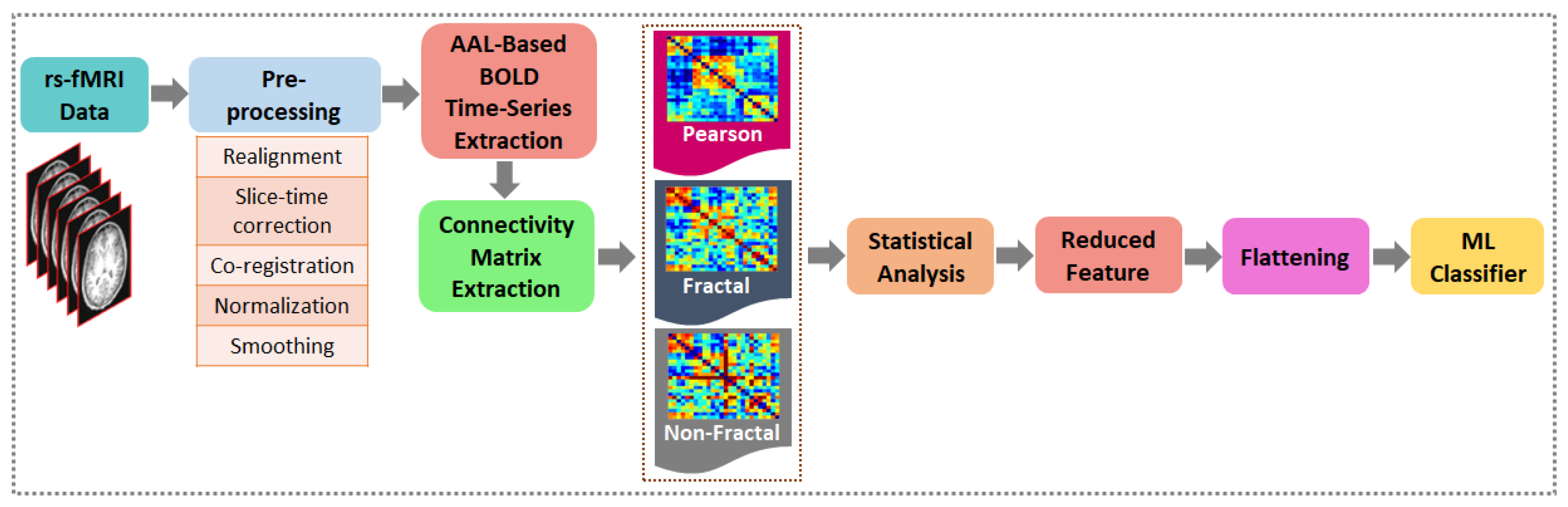
3. Methodology
3.1. Description of Dataset
3.2. Data Pre-Processing
3.3. BOLD Time-Series Signals Extraction
3.4. Functional Connectivity of rs-fMRI Signals
3.4.1. Pearson Correlation Coefficient
3.4.2. Wavelet Analysis for Fractal Connectivity
3.4.3. Wavelet Analysis for Nonfractal Connectivity
3.5. Statistical Analysis, Feature Reduction and Flattening of Functional Connections
3.6. ML Classifier
3.7. Performance Evaluation
4. Result and Discussion
4.1. Statistical Analysis
4.2. Classification of AD vs. NC Based on Nonfractal Connectivity
4.2.1. Selection of the Best ML Classifier
4.2.2. Evaluation of Significant Functional Connections
4.2.3. Classification of AD vs. NC Using SVM
4.3. Investigation on the Proposed AD Classification Using XHSLF+ADNI Dataset
4.4. Comparison with Related Works
5. Conclusions
Author Contributions
Funding
- Fundamental Research Grant Scheme (FRGS/1/2021/TK0/UTP/02/17), awarded by the Ministry of Higher Education (MOHE), Malaysia.
- Higher Institutional Centre of Excellence (HICoE) Grant awarded to the Centre for Intelligent Signal and Imaging Research (CISIR), by the Ministry of Higher Education (MOHE), Malaysia.
- YUTP-Fundamental Research Grant (YUTP-FRG 015LC0-292), awarded by the Yayasan Universiti Teknologi PETRONAS (YUTP).
Conflicts of Interest
Abbreviations
| AAL | Automated Anatomical Labeling |
| AD | Alzheimer’s Disease |
| ADNI | Alzheimer’s Disease Neuroimaging Initiative |
| ANOVA | Analysis of Variance |
| ARIMA | Auto Regressive Fractionally Integrated Moving Average |
| ARMA | Auto Regressive Moving Average |
| BOLD | Blood Oxygen Level Dependent |
| CDR | Clinical Dementia Rating |
| CONN | Connectivity toolbox |
| CSF | Cerebrospinal Fluid |
| DPABI | Data Processing & Analysis for Brain Imaging |
| DWT | Discrete Wavelet Transform |
| EEG | Electroencephalography |
| FGN | Fractional Gaussain Noise |
| FIN | Fractionally Integrated Noise |
| FIP | Fractal Integrated Process |
| fMRI | Functional Magnetic Resonance Imaging |
| LM | Long Memory |
| MCI | Mild Cognitive Impairment |
| ML | Machine Learning |
| MMSE | Mini Mental State Examination |
| NC | Normal Controls |
| PET | Positron Emission Tomography |
| SLF | Santa Lucia Foundation |
| sMRI | Structural Magnetic Resonance Imaging |
| XH | Xuanwu Hospital |
| XHSLF | Xuanwu Hospital Santa Lucia Foundation |
References
- Delbeuck, X.; Van der Linden, M.; Collette, F. Alzheimer’ Disease as a Disconnection Syndrome? Neuropsychol. Rev. 2003, 13, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Stam, C.; Jones, B.; Nolte, G.; Breakspear, M.; Scheltens, P. Small-World Networks and Functional Connectivity in Alzheimer’s Disease. Cerebral Cortex 2006, 17, 92–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Supekar, K.; Menon, V.; Rubin, D.; Musen, M.; Greicius, M.D. Network Analysis of Intrinsic Functional Brain Connectivity in Alzheimer’s Disease. PLoS Comput. Biol. 2008, 4, e1000100. [Google Scholar] [CrossRef]
- Feng, Q.; Wang, M.; Song, Q.; Wu, Z.; Jiang, H.; Pang, P.; Liao, Z.; Yu, E.; Ding, Z. Correlation Between Hippocampus MRI Radiomic Features and Resting-State Intrahippocampal Functional Connectivity in Alzheimer’s Disease. Front. Neurosci. 2019, 13, 435. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, F.; Zulifqar, H.; Malik, T. Classification of Alzheimer disease among susceptible brain regions. Int. J. Imaging Syst. Technol. 2019, 29, 222–233. [Google Scholar] [CrossRef]
- De Souza Duran, J.M.R.L.K.F.F.L.; Kubo, R.; Ono, C.R.; Leite, C.C.; Smid, J.; Nitrini, R.; Buchpiguel, C.A.; Busatto, G.F. Selecting the most relevant brain regions to discriminate Alzheimer’s disease patients from healthy controls using multiple kernel learning: A comparison across functional and structural imaging modalities and atlases. Neuroimage Clin. 2018, 17, 628–641. [Google Scholar] [CrossRef]
- Sun, L.; Patel, R.; Liu, J.; Chen, K.; Wu, T.; Li, J.; Reiman, E.; Ye, J. Mining Brain Region Connectivity for Alzheimer’s Disease Study via Sparse Inverse Covariance Estimation. In Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Paris, France, 28 June–1 July 2009; pp. 1335–1344. [Google Scholar] [CrossRef]
- Pei, S.; Guan, J.; Zhou, S. Classifying early and late mild cognitive impairment stages of Alzheimer’s disease by fusing default mode networks extracted with multiple seeds. BMC Bioinform. 2018, 19, 523. [Google Scholar] [CrossRef]
- Westman, E.; Muehlboeck, J.S.; Simmons, A. Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. NeuroImage 2012, 62, 229–238. [Google Scholar] [CrossRef]
- Hansson, O.; Lehmann, S.; Otto, M.; Zetterberg, H.; Lewczuk, P. Advantages and disadvantages of the use of the CSF Amyloid β (Aβ) 42/40 ratio in the diagnosis of Alzheimer’s Disease. Alzheimer’s Res. Ther. 2019, 11, 34. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Teixeira, J.P.; Garrett, C.; Alves, D.; Freitas, D. Alzheimer’s Early Prediction with Electroencephalogram. Procedia Comput. Sci. 2016, 100, 865–871. [Google Scholar] [CrossRef] [Green Version]
- Houmani, N.; Vialatte, F.; Gallego-Jutglà, E.; Dreyfus, G.; Nguyen-Michel, V.H.; Mariani, J.; Kinugawa, K. Diagnosis of Alzheimer’s disease with Electroencephalography in a differential framework. PLoS ONE 2018, 13, e0193607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhang, J.; Wei, X.; Zhang, Q. Alzheimer’s Disease Classification Using Structural MRI Based on Convolutional Neural Networks. In Proceedings of the 2020 2nd International Conference on Big-Data Service and Intelligent Computation, Xiamen, China, 3–5 December 2020; pp. 7–13. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, Y.; Zhu, D.C.; Bozoki, A.C.; Li, T. Classification of Alzheimer’s Disease, Mild Cognitive Impairment and Normal Control Subjects Using Resting-State fMRI Based Network Connectivity Analysis. IEEE J. Transl. Eng. Health Med. 2018, 6, 1801009. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Yao, Z.; Li, Y.; Zhang, Y.; Hu, B.; Wu, D.; Alzheimer’s Disease Neuroimaging Initiative. Brain Connectivity Based Prediction of Alzheimer’s Disease in Patients With Mild Cognitive Impairment Based on Multi-Modal Images. Front. Hum. Neurosci. 2019, 13, 399. [Google Scholar] [CrossRef] [PubMed]
- Suk, H.; Lee, S.; Shen, D. Deep ensemble learning of sparse regression models for brain disease diagnosis. Med. Image Anal. 2017, 37, 101–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esmaeilzadeh, S.; Belivanis, D.; Pohl, K.; Adeli, E. End-To-End Alzheimer’s Disease Diagnosis and Biomarker Identification. Mach. Learn. Med. Imaging MLMI 2018, 11046, 337–345. [Google Scholar]
- Albright, J. Forecasting the progression of Alzheimer’s disease using neural networks and a novel preprocessing algorithm. Alzheimer’s Dement. 2019, 5, 483–491. [Google Scholar] [CrossRef]
- Castellazzi, G.; Cuzzoni, M.G.; Cotta Ramusino, M.; Martinelli, D.; Denaro, F.; Ricciardi, A.; Vitali, P.; Anzalone, N.; Bernini, S.; Palesi, F.; et al. A Machine Learning Approach for the Differential Diagnosis of Alzheimer and Vascular Dementia Fed by MRI Selected Features. Front. Neuroinform. 2020, 14, 25. [Google Scholar] [CrossRef]
- Noroozi, A.; Rezghi, M. A Tensor-Based Framework for rs-fMRI Classification and Functional Connectivity Construction. Front. Neuroinform. 2020, 14, 46. [Google Scholar] [CrossRef]
- De Vos, F.; Koini, M.; Schouten, T.M.; Seiler, S.; van der Grond, J.; Lechner, A.; Schmidt, R.; de Rooij, M.; Rombouts, S.A. A comprehensive analysis of resting state fMRI measures to classify individual patients with Alzheimer’s disease. Neuroimage 2018, 167, 62–72. [Google Scholar] [CrossRef] [Green Version]
- Ju, R.; Hu, C.; Zhou, P.; Li, Q. Early Diagnosis of Alzheimer’s Disease Based on Resting-State Brain Networks and Deep Learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 244–257. [Google Scholar] [CrossRef]
- Wang, B.; Niu, Y.; Miao, L.; Cao, R.; Yan, P.; Guo, H.; Li, D.; Guo, Y.; Yan, T.; Wu, J.; et al. Decreased Complexity in Alzheimer’s Disease: Resting-State fMRI Evidence of Brain Entropy Mapping. Front. Aging Neurosci. 2017, 9, 378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Kong, R.; Liégeois, R.; Orban, C.; Tan, Y.; Sun, N.; Holmes, A.J.; Sabuncu, M.R.; Ge, T.; Yeo, B.T. Global signal regression strengthens association between resting-state functional connectivity and behavior. NeuroImage 2019, 196, 126–141. [Google Scholar] [CrossRef] [PubMed]
- Birn, R.M.; Diamond, J.B.; Smith, M.A.; Bandettini, P.A. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. NeuroImage 2006, 31, 1536–1548. [Google Scholar] [CrossRef] [PubMed]
- Wink, A.M.; Bullmore, E.; Barnes, A.; Bernard, F.; Suckling, J. Monofractal and multifractal dynamics of low frequency endogenous brain oscillations in functional MRI. Hum. Brain Mapp. 2008, 29, 791–801. [Google Scholar] [CrossRef]
- Jang, H.; Plis, S.M.; Calhoun, V.D.; Lee, J.H. Task-specific feature extraction and classification of fMRI volumes using a deep neural network initialized with a deep belief network: Evaluation using sensorimotor tasks. NeuroImage 2017, 145, 314–328. [Google Scholar] [CrossRef] [Green Version]
- Zarahn, E.; Aguirre, G.; D’Esposito, M. Empirical Analyses of BOLD fMRI Statistics. NeuroImage 1997, 5, 179–197. [Google Scholar] [CrossRef]
- Stam, C.J.; de Bruin, E.A. Scale-free dynamics of global functional connectivity in the human brain. Hum. Brain Mapp. 2004, 22, 97–109. [Google Scholar] [CrossRef]
- Maxim, V.; Şendur, L.; Fadili, J.; Suckling, J.; Gould, R.; Howard, R.; Bullmore, E. Fractional Gaussian noise, functional MRI and Alzheimer’s disease. Neuroimage 2005, 25, 141–158. [Google Scholar] [CrossRef] [Green Version]
- Eke, A.; Hermán, P.; Hajnal, M. Fractal and Noisy CBV Dynamics in Humans: Influence of Age and Gender. J. Cereb. Blood Flow Metab. 2006, 26, 891–898. [Google Scholar] [CrossRef]
- Peter Herman, A.E. Nonlinear Analysis of Blood Cell Flux Fluctuations in the Rat Brain Cortex during Stepwise Hypotension Challenge. J. Cereb. Blood Flow Metab. 2006, 26, 1189–1197. [Google Scholar] [CrossRef]
- Van de Ville, D.; Britz, J.; Michel, C.M. EEG microstate sequences in healthy humans at rest reveal scale-free dynamics. Proc. Natl. Acad. Sci. USA 2010, 107, 18179–18184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Expert, P.; Lambiotte, R.; Chialvo, D.R.; Christensen, K.; Jensen, H.J.; Sharp, D.J.; Turkheimer, F. Self-similar correlation function in brain resting-state functional magnetic resonance imaging. J. R. Soc. Interface 2011, 8, 472–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, W.; Achard, S.; Stadler, J.; Brückner, B.; Seiffert, U. Fractal analysis of resting state functional connectivity of the brain. In Proceedings of the 2012 International Joint Conference on Neural Networks (IJCNN), Brisbane, QLD, Australia, 10–15 June 2012; pp. 1–8. [Google Scholar] [CrossRef] [Green Version]
- West, B.J.; Zhang, R.; Sanders, A.W.; Miniyar, S.; Zuckerman, J.H.; Levine, B.D. Fractal fluctuations in cardiac time series. Phys. A Stat. Mech. Its Appl. 1999, 270, 552–566. [Google Scholar] [CrossRef]
- Govindpani, K.; McNamara, L.G.; Smith, N.R.; Vinnakota, C.; Waldvogel, H.J.; Faull, R.L.; Kwakowsky, A. Vascular Dysfunction in Alzheimer’s Disease: A Prelude to the Pathological Process or a Consequence of It? J. Clin. Med. 2019, 8, 651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achard, S.; Bassett, D.S.; Meyer-Lindenberg, A.; Bullmore, E. Fractal connectivity of long-memory networks. Phys. Rev. E 2008, 77, 036104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, W.; Limperopoulos, C. Study on Linear Combination of Long Memory Processes Corrupted by Additive Noises for fMRI Time Series Analysis. bioRxiv 2017. [Google Scholar] [CrossRef] [Green Version]
- Karasu, S.; Altan, A.; Bekiros, S.; Ahmad, W. A new forecasting model with wrapper-based feature selection approach using multi-objective optimization technique for chaotic crude oil time series. Energy 2020, 212, 118750. [Google Scholar] [CrossRef]
- Karasu, S.; Altan, A. Crude oil time series prediction model based on LSTM network with chaotic Henry gas solubility optimization. Energy 2022, 242, 122964. [Google Scholar] [CrossRef]
- Rodríguez-Liñares, L.; Méndez, A.J.; Lado, M.J.; Oliv, D.N.; Vila, X.A.; Gómez-Conde, I. An open source tool for heart rate variability spectral analysis. Comput. Methods Programs Biomed. 2011, 103, 39–50. [Google Scholar] [CrossRef]
- Wendt, H.; Abry, P. Multifractality Tests Using Bootstrapped Wavelet Leaders. IEEE Trans. Signal Process. 2007, 55, 4811–4820. [Google Scholar] [CrossRef] [Green Version]
- Jaffard, S.; Lashermes, B.; Abry, P. Wavelet Leaders in Multifractal Analysis. In Wavelet Analysis and Applications; Qian, T., Vai, M.I., Xu, Y., Eds.; Birkhäuser Basel: Basel, Switzerland, 2007; pp. 201–246. [Google Scholar]
- Liu, Y.; Yu, C.; Zhang, X.; Liu, J.; Duan, Y.; Alexander-Bloch, A.F.; Liu, B.; Jiang, T.; Bullmore, E. Impaired Long Distance Functional Connectivity and Weighted Network Architecture in Alzheimer’s Disease. Cereb. Cortex 2013, 24, 1422–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascali, D.; DiNuzzo, M.; Gili, T.; Moraschi, M.; Fratini, M.; Maraviglia, B.; Serra, L.; Bozzali, M.; Giove, F. Intrinsic Patterns of Coupling between Correlation and Amplitude of Low-Frequency fMRI Fluctuations Are Disrupted in Degenerative Dementia Mainly due to Functional Disconnection. PLoS ONE 2015, 10, e0120988. [Google Scholar] [CrossRef] [PubMed]
- Petersen, R.C.; Aisen, P.S.; Beckett, L.A.; Donohue, M.C.; Gamst, A.C.; Harvey, D.J.; Jack, C.R., Jr.; Jagust, W.J.; Shaw, L.M.; Toga, A.W.; et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): Clinical characterization. Neurology 2010, 74, 201–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, C.G.; Wang, X.D.; Zuo, X.N.; Zang, Y.F. DPABI: Data Processing & Analysis for (Resting-State) Brain Imaging. Neuroinformatics 2016, 14, 339–351. [Google Scholar] [CrossRef]
- Kirch, W. (Ed.) Pearson’s Correlation Coefficient. In Encyclopedia of Public Health; Springer: Berlin/Heidelberg, Germany, 2008; pp. 1090–1091. [Google Scholar] [CrossRef]
- Percival, D.B.; Walden, A.T. Frontmatter. In Wavelet Methods for Time Series Analysis; Cambridge Series in Statistical and Probabilistic Mathematics; Cambridge University Press: New York, NY, USA, 2000; pp. 1–6. [Google Scholar]
- Xu, L.; Liang, G.; Liao, C.; Chen, G.D.; Chang, C.C. An Efficient Classifier for Alzheimer’s Disease Genes Identification. Molecules 2018, 23, 3140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasani, P.H.; Kasani, S.H.; Kim, Y.; Yun, C.H.; Choi, S.H.; Jang, J.W. An Evaluation of Machine Learning Classifiers for Prediction of Alzheimer’s Disease, Mild Cognitive Impairment and Normal Cognition. In Proceedings of the 2021 International Conference on Information and Communication Technology Convergence (ICTC), Jeju Island, Korea, 20–22 October 2021; pp. 362–367. [Google Scholar] [CrossRef]
- Zhu, Q.; Wang, Y.; Zhuo, C.; Xu, Q.; Yao, Y.; Liu, Z.; Li, Y.; Sun, Z.; Wang, J.; Lv, M.; et al. Classification of Alzheimer’s Disease Based on Abnormal Hippocampal Functional Connectivity and Machine Learning. Front. Aging Neurosci. 2022, 14. [Google Scholar] [CrossRef] [PubMed]





| XH (21) + SLF (10) | XH | ADNI | ||
|---|---|---|---|---|
| NC | AD | NC | AD | |
| No. of Subject | 31 | 35 | 30 | 30 |
| Gender (M/F) | 14/17 | 17/18 | 13/17 | 15/15 |
| Age (Year) | 63 ± 8.85 | 65.8 ± 8.3 | 75.25 ± 6.9 | 72.81 ± 7.17 |
| MMSE | 28.9 ± 1.035 | 10.1 ± 6.7 | 24–30 | 20–26 |
| CDR | 0 | 1–3 | 0 | 0.5–1 |
| Acquisition Protocol | |||
|---|---|---|---|
| XH | SLF | ADNI | |
| Field Strength | 3.0 Tesla | 3.0 Tesla | 3.0 Tesla |
| Flip Angle | 90.0 degrees | 70.0 degrees | 80.0 degrees |
| Manufacturer | Siemens | Siemens | Philips |
| Matrix X | 64.0 pixels | 64.0 pixels | 64.0 pixels |
| Matrix Y | 64.0 pixels | 64.0 pixels | 64.0 pixels |
| Slices | 5440 | 7040 | 6720 |
| Time points | 170 | 220 | 140 |
| TR | 2000.0 ms | 2080.0 ms | 3000.0 ms |
| TE | 30.0 ms | 30.0 ms | 30.0 ms |
| FOV | 220 mm × 220 mm | 256 mm × 224 mm | 212 mm × 212 mm |
| Slice Thickness | 3 mm | 2.5 mm | 3.3 mm |
| Classifier | Nonfractal | Fractal | Pearson Correlation | |||
|---|---|---|---|---|---|---|
| XHSLF | ADNI | XHSLF | ADNI | XHSLF | ADNI | |
| KNN (Fine) | 80.64 ± 2.51 | 72.34 ± 3.66 | 79.96 ± 0.78 | 58.83 ± 2.69 | 63.22 ± 1.19 | 63.32 ± 2.58 |
| Decision tree (Fine) | 63.72 ± 4.27 | 59.51 ± 3.41 | 60.66± 5.05 | 54.66 ± 6.33 | 60.98 ± 4.25 | 54.02 ± 5.13 |
| Bagged trees (Fine) | 71.78 ± 4.33 | 62.98 ± 0.64 | 70.97 ± 2.51 | 62.52 ± 1.32 | 62.58 ± 2.65 | 62.82 ± 0.733 |
| SVM (Linear) | 90.3 ± 1.23 | 83.3 ± 0.67 | 82.3 ± 0.51 | 71.67 ± 3.15 | 72.6 ± 0.71 | 70 ± 0.76 |
| Dataset | Nonfractal | Fractal | Pearson Correlation |
|---|---|---|---|
| XHSLF | 90.3 (0.05) | 72.6 (0.007) | 51.6 (0.009) |
| ADNI | 83.3 (0.05) | 63.3 (0.012) | 68.3 (0.0006) |
| Evaluation Metric | Nonfractal | Fractal | Pearson Correlation | |||
|---|---|---|---|---|---|---|
| XHSLF | ADNI | XHSLF | ADNI | XHSLF | ADNI | |
| Sensitivity | 87.87 | 100 | 77.77 | 70.96 | 68.42 | 71.42 |
| Specificity | 93.1 | 75 | 88.46 | 72.41 | 79.16 | 68.75 |
| Accuracy | 90.3 | 83.3 | 82.3 | 71.6 | 72.6 | 70 |
| Precision | 93.54 | 66.66 | 90.3 | 73.3 | 83.87 | 66.66 |
| FPR | 6.89 | 25 | 11.5 | 27.5 | 20.83 | 31.25 |
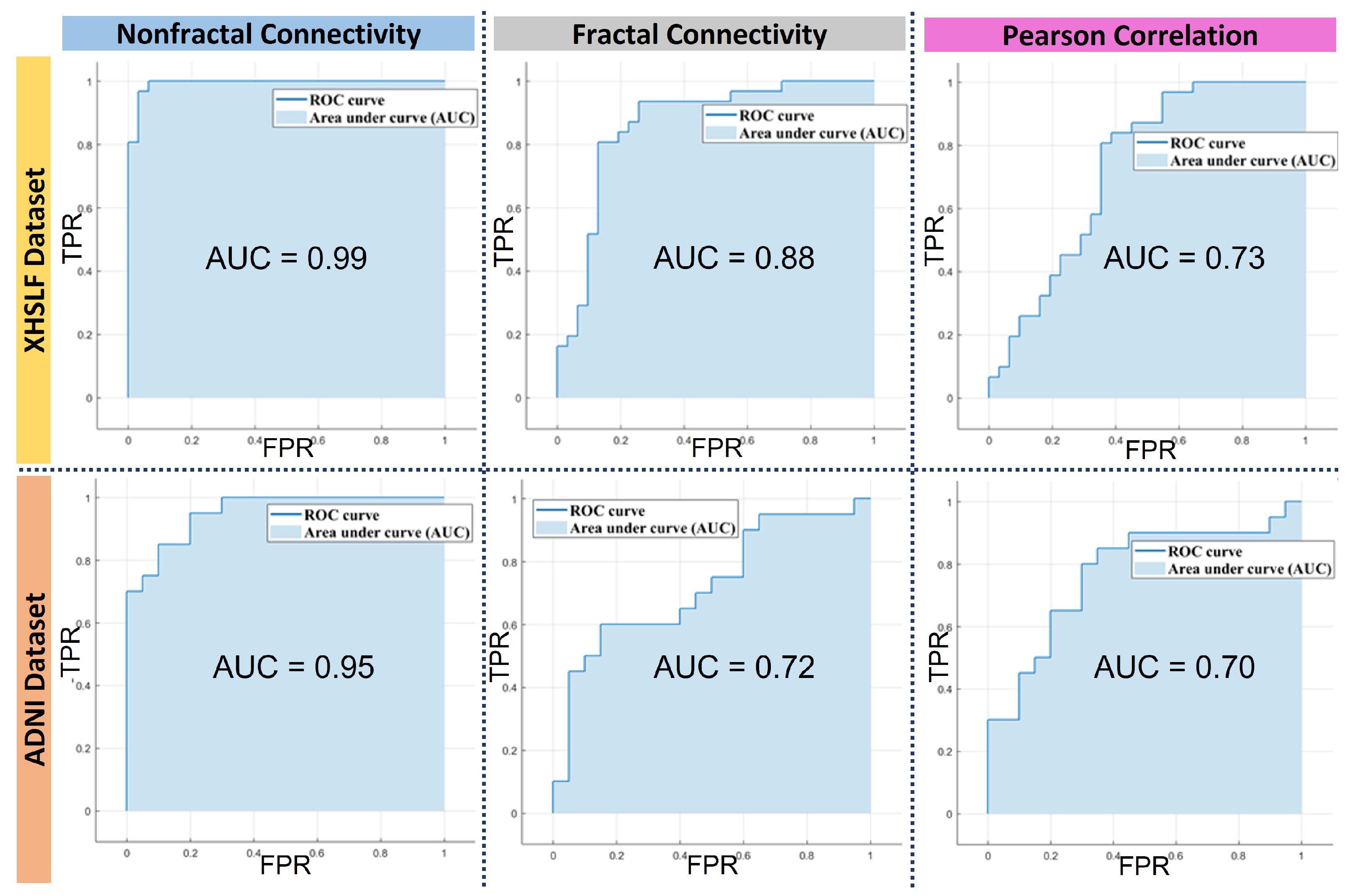
| AUC | 0.98 | 0.93 | 0.88 | 0.72 | 0.73 | 0.70 |
| Classifier | Nonfractal | Fractal | Pearson Correlation |
|---|---|---|---|
| XHSLF+ADNI | XHSLF+ADNI | XHSLF+ADNI | |
| KNN (Fine) | 90.2 ± 0.02 | 73.8 ± 1.85 | 60.26 ± 1.13 |
| Decision tree (Fine) | 59.8 ± 2.75 | 58.13 ± 1.17 | 58.19 ± 2.99 |
| Bagged trees | 67.2 ± 0.99 | 65.93 ± 2.58 | 64.84 ± 1.88 |
| SVM (linear) | 82.8 ± 0.71 | 66.49 ± 1.42 | 73 ± 1.45 |
| Evaluation Metric | Nonfractal | Fractal | Pearson Correlation |
|---|---|---|---|
| XHSLF+ADNI | XHSLF+ADNI | XHSLF+ADNI | |
| Sensitivity | 91.5 | 75.4 | 70.5 |
| Specificity | 88.88 | 72.3 | 75.9 |
| Accuracy | 90.2 | 73.8 | 73 |
| Precision | 88.52 | 70.49 | 78.6 |
| FPR | 11.11 | 27.69 | 24.07 |
| AUC | 0.9 | 0.74 | 0.77 |
| Method | Features | Dataset (Number of Subjects) | Evaluation Metric | ||||
|---|---|---|---|---|---|---|---|
| NC | AD | Accuracy | Sensitivity | Specificity | AUC | ||
| De Vos et al. (2018) | Functional connectivity | 173 | 77 | 79 | 86 | 71 | 0.85 |
| Kasani et al. (2021) | Correlation connectivity | 173 | 74 | 82.75 | 82.75 | - | - |
| Zhu et al. (2022) | Functional connectivity | 45 | 44 | 82.02 | - | - | - |
| Our Proposed Method (XHSLF) | Nonfractal connectivity | 31 | 31 | 90.3 | 87.87 | 93.1 | 0.98 |
| Our Proposed Method (ADNI) | Nonfractal connectivity | 30 | 30 | 83.3 | 100 | 75 | 0.93 |
| Our Proposed Method (XHSLF+ADNI) | Nonfractal connectivity | 61 | 61 | 90.2 | 91.5 | 88.88 | 0.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadiq, A.; Yahya, N.; Tang, T.B.; Hashim, H.; Naseem, I. Wavelet-Based Fractal Analysis of rs-fMRI for Classification of Alzheimer’s Disease. Sensors 2022, 22, 3102. https://doi.org/10.3390/s22093102
Sadiq A, Yahya N, Tang TB, Hashim H, Naseem I. Wavelet-Based Fractal Analysis of rs-fMRI for Classification of Alzheimer’s Disease. Sensors. 2022; 22(9):3102. https://doi.org/10.3390/s22093102
Chicago/Turabian StyleSadiq, Alishba, Norashikin Yahya, Tong Boon Tang, Hilwati Hashim, and Imran Naseem. 2022. "Wavelet-Based Fractal Analysis of rs-fMRI for Classification of Alzheimer’s Disease" Sensors 22, no. 9: 3102. https://doi.org/10.3390/s22093102







