A Novel Fluorescent Sensor Based on Aptamer and qPCR for Determination of Glyphosate in Tap Water
Abstract
:1. Introduction
2. Experiment
2.1. Materials and Instruments
2.2. Magnetic Bead Activation
2.3. Preparation of Mbs@dsDNA and Pre-Experiment
2.4. Specificity Detection
2.5. Determination of Glyphosate
2.6. Actual Sample Testing
3. Results and Discussion
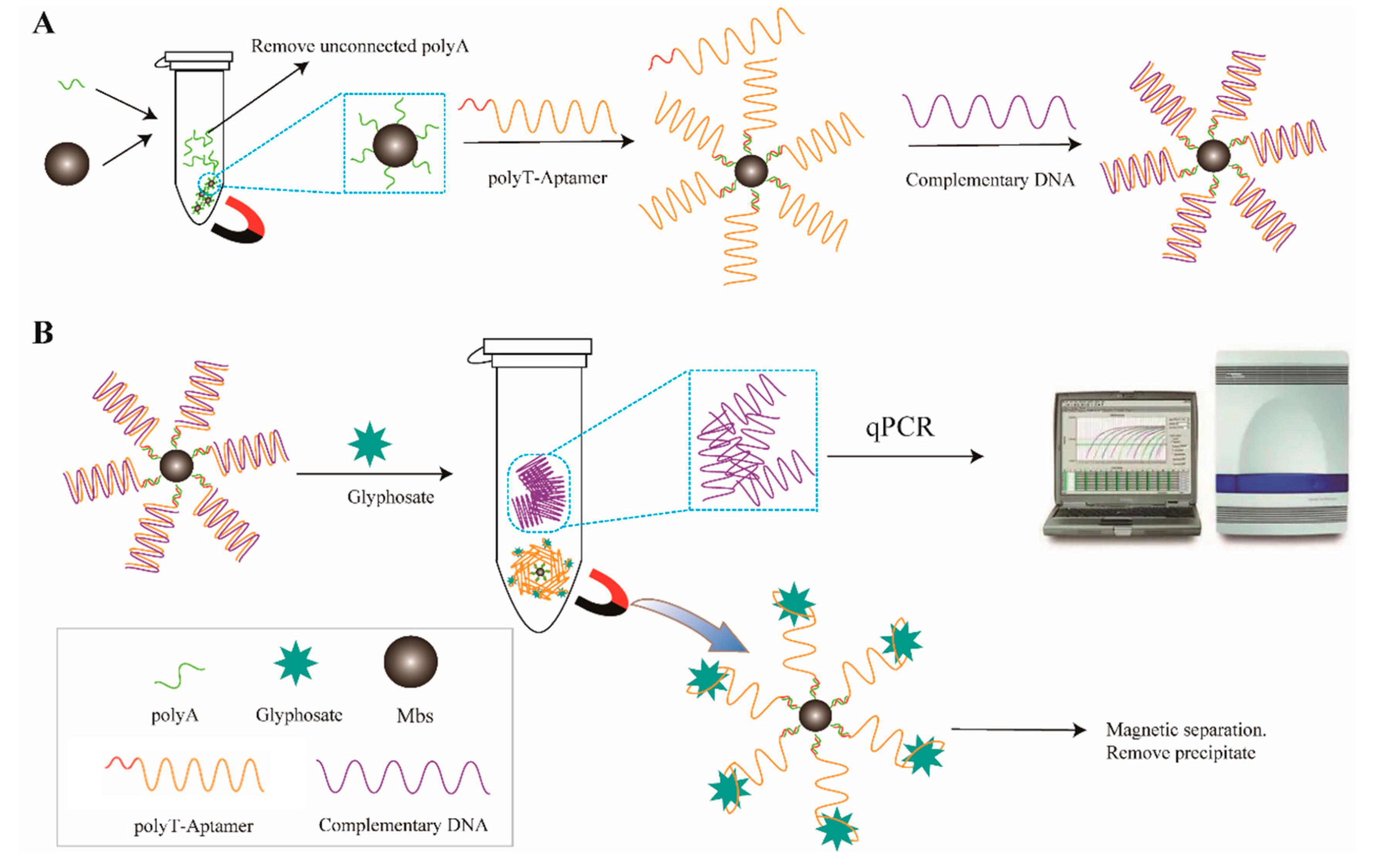
3.1. Experimental Principle
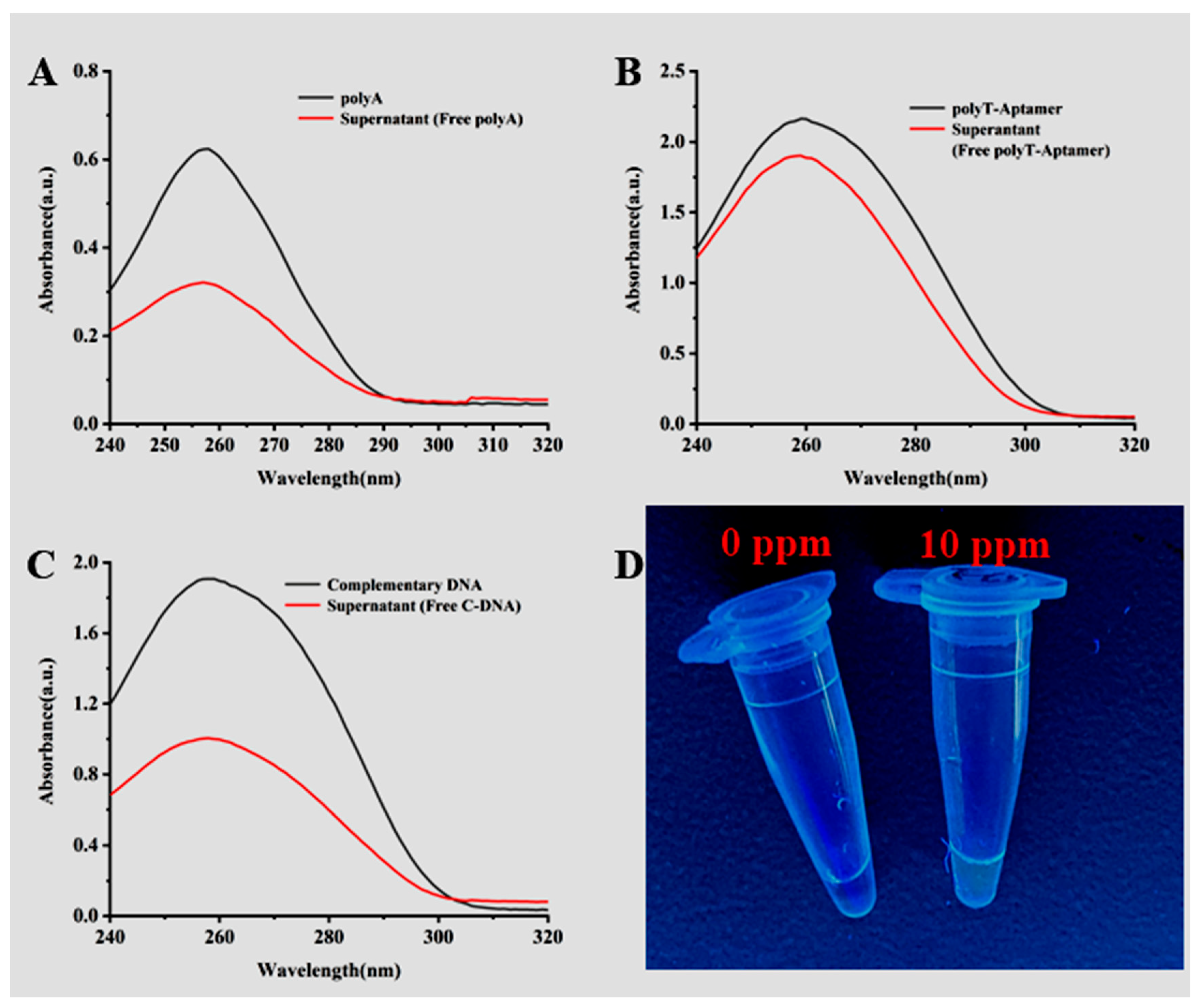
3.2. Characterization of Mbs@dsDNA and Aptamer Sensor Feasibility Validation
3.3. Validation of the Specificity of the Aptamer Sensor
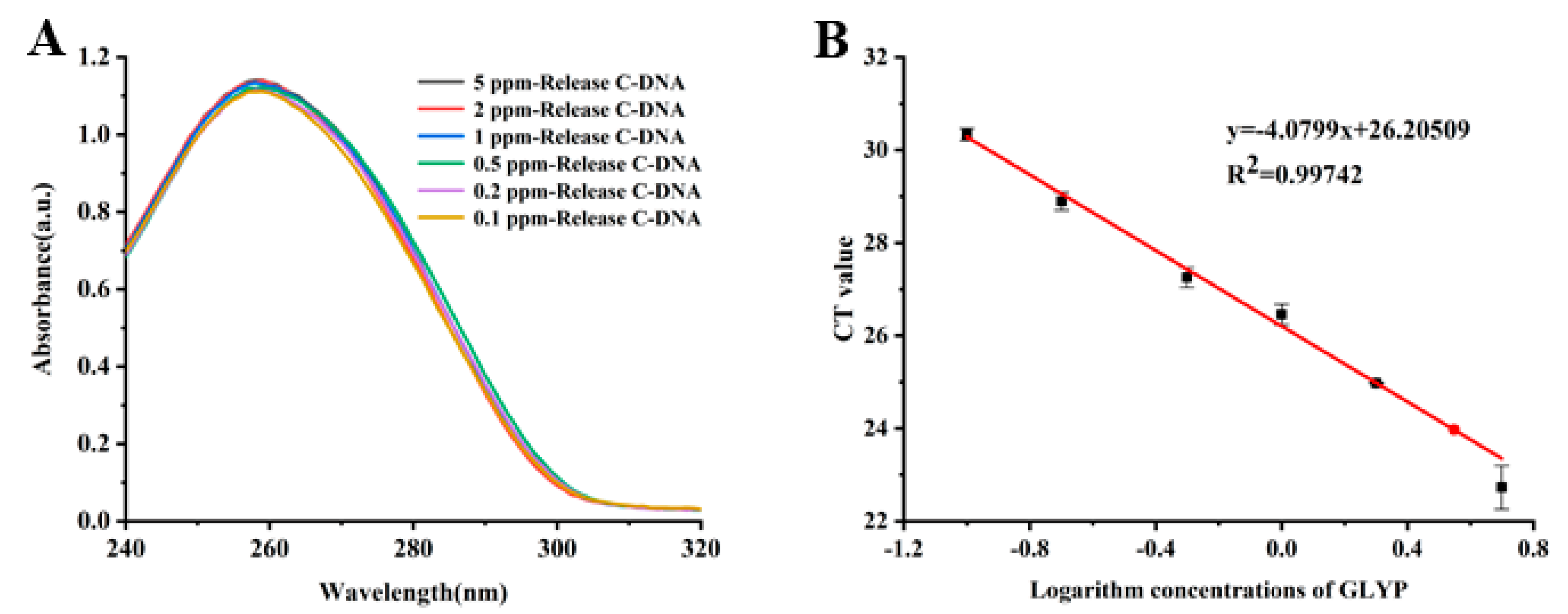
3.4. Glyphosate Detection and qPCRs Based on Mbs@dsDNA
3.5. Application of This and Other Methods to the Detection of Glyphosate in Real Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Benbrook, C.M. Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 2016, 28, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, J.P.K.; Sethi, N.; Mohan, A. Analysis of the glyphosate herbicide in water, soil and food using derivatising agents. Environ. Chem. Lett. 2017, 15, 85–100. [Google Scholar] [CrossRef]
- Jimenez-Lopez, J.; Llorent-Martinez, E.J.; Ortega-Barrales, P.; Ruiz-Medina, A. Graphene quantum dots-silver nanoparticles as a novel sensitive and selective luminescence probe for the detection of glyphosate in food samples. Talanta 2020, 207, 120344. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, L.; Sharma, A.S.; Chen, M.; Chen, Q. A system composed of polyethylenimine-capped upconversion nanoparticles, copper (II), hydrogen peroxide and 3,3’,5,5’-tetramethylbenzidine for colorimetric and fluorometric determination of glyphosate. Mikrochim. Acta 2019, 186, 835. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Lin, Y.S.; Xiao, G.T.; Chiu, T.C.; Hu, C.C. A highly selective and sensitive nanosensor for the detection of glyphosate. Talanta 2016, 161, 94–98. [Google Scholar] [CrossRef]
- Duke, S.O.; Powles, S.B. Glyphosate: A once-in-a-century herbicide. Pest Manag. Sci. 2008, 64, 319–325. [Google Scholar] [CrossRef]
- Prasad, B.B.; Jauhari, D.; Tiwari, M.P. Doubly imprinted polymer nanofilm-modified electrochemical sensor for ultra-trace simultaneous analysis of glyphosate and glufosinate. Biosens. Bioelectron. 2014, 59, 81–88. [Google Scholar] [CrossRef]
- Regiart, M.; Kumar, A.; Goncalves, J.M.; Silva, G.J.; Masini, J.C.; Angnes, L.; Bertotti, M. An Electrochemically Synthesized Nanoporous Copper Microsensor for Highly Sensitive and Selective Determination of Glyphosate. Chemelectrochem 2020, 7, 1558–1566. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Datta, S.; Wani, A.B.; Dhanjal, D.S.; Romero, R.; Singh, J. Glyphosate uptake, translocation, resistance emergence in crops, analytical monitoring, toxicity and degradation: A review. Environ. Chem. Lett. 2020, 18, 663–702. [Google Scholar] [CrossRef]
- Bringolf, R.B.; Cope, W.G.; Mosher, S.; Barnhart, M.C.; Shea, D. Acute and chronic toxicity of glyphosate compounds to glochidia and juveniles of Lampsilis siliquoidea (Unionidae). Environ. Toxicol. Chem. 2007, 26, 2094–2100. [Google Scholar] [CrossRef]
- Xu, J.; Smith, S.; Smith, G.; Wang, W.; Li, Y. Glyphosate contamination in grains and foods: An overview. Food Control 2019, 106, 106710. [Google Scholar] [CrossRef]
- Araujo, A.S.; Monteiro, R.T.; Abarkeli, R.B. Effect of glyphosate on the microbial activity of two Brazilian soils. Chemosphere 2003, 52, 799–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khrolenko, M.V.; Wieczorek, P.P. Determination of glyphosate and its metabolite aminomethylphosphonic acid in fruit juices using supported-liquid membrane preconcentration method with high-performance liquid chromatography and UV detection after derivatization with p-toluenesulphonyl chloride. J. Chromatogr. A 2005, 1093, 111–117. [Google Scholar] [PubMed]
- Connolly, A.; Koslitz, S.; Bury, D.; Bruning, T.; Conrad, A.; Kolossa-Gehring, M.; Coggins, M.A.; Koch, H.M. Sensitive and selective quantification of glyphosate and aminomethylphosphonic acid (AMPA) in urine of the general population by gas chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2020, 1158, 122348. [Google Scholar] [CrossRef] [PubMed]
- Kudzin, Z.H.; Gralak, D.K.; Drabowicz, J.; Luczak, J. Novel approach for the simultaneous analysis of glyphosate and its metabolites. J. Chromatogr. A 2002, 947, 129–141. [Google Scholar] [CrossRef]
- Morgan, M.A.; Griffith, C.M.; Dinges, M.M.; Lyon, Y.A.; Julian, R.R.; Larive, C.K. Evaluating sub-lethal stress from Roundup® exposure in Artemia franciscana using (1)H NMR and GC-MS. Aquat. Toxicol. 2019, 212, 77–87. [Google Scholar] [CrossRef]
- Bressan, I.G.; Llesuy, S.F.; Rodriguez, C.; Ferloni, A.; Dawidowski, A.R.; Figar, S.B.; Gimenez, M.I. Optimization and validation of a liquid chromatography-tandem mass spectrometry method for the determination of glyphosate in human urine after pre-column derivatization with 9-fluorenylmethoxycarbonyl chloride. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2021, 1171, 122616. [Google Scholar] [CrossRef]
- Gormez, E.; Golge, O.; Kabak, B. Quantification of fosetyl-aluminium /phosphonic acid and other highly polar residues in pomegranates using Quick Polar Pesticides method involving liquid chromatography-tandem mass spectrometry measurement. J. Chromatogr. A 2021, 1642, 462038. [Google Scholar] [CrossRef]
- Jost, U.; Habedank, F. Two-dimensional hydrophilic interaction and reversed phase liquid chromatography easily extracted pesticides and polar pesticides multi-residue method—A concept. J. Chromatogr. A 2020, 1621, 461040. [Google Scholar] [CrossRef]
- Thompson, T.S.; van den Heever, J.P.; Limanowka, R.E. Determination of glyphosate, AMPA, and glufosinate in honey by online solid-phase extraction-liquid chromatography-tandem mass spectrometry. Food Addit. Contam. Part A 2019, 36, 434–446. [Google Scholar] [CrossRef]
- Geerdink, R.B.; Hassing, M.; Ayarza, N.; Bruggink, C.; Wielheesen, M.; Claassen, J.; Epema, O.J. Analysis of glyphosate, AMPA, Glufosinate and MPPA with ION chromatography tandem mass spectrometry using A membrane suppressor in the ammonium form application to surface water of low to moderate salinity. Anal. Chim. Acta 2020, 1133, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Schutze, A.; Morales-Agudelo, P.; Vidal, M.; Calafat, A.M.; Ospina, M. Quantification of glyphosate and other organophosphorus compounds in human urine via ion chromatography isotope dilution tandem mass spectrometry. Chemosphere 2021, 274, 129427. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.W.; Deng, T.; Liang, S.L.; Tan, X.F.; Meng, J.X. Determination of Herbicides and Its Metabolite in Soil and Water Samples by Capillary Electrophoresis-laser Induced Fluorescence Detection Using Microwave-assisted Derivatization. Anal. Sci. 2014, 30, 759–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wimmer, B.; Pattky, M.; Zada, L.G.; Meixner, M.; Haderlein, S.B.; Zimmermann, H.P.; Huhn, C. Capillary electrophoresis-mass spectrometry for the direct analysis of glyphosate: Method development and application to beer beverages and environmental studies. Anal. Bioanal. Chem. 2020, 412, 4967–4983. [Google Scholar] [CrossRef] [PubMed]
- Byer, J.D.; Struger, J.; Klawunn, P.; Todd, A.; Sverko, E. Low cost monitoring of glyphosate in surface waters using the ELISA method: An evaluation. Environ. Sci. Technol. 2008, 42, 6052–6057. [Google Scholar] [CrossRef]
- El-Gendy, K.; Mosallam, E.; Ahmed, N.; Aly, N. Determination of glyphosate residues in Egyptian soil samples. Anal. Biochem. 2018, 557, 1–6. [Google Scholar] [CrossRef]
- Guan, J.; Yang, J.; Zhang, Y.; Zhang, X.; Deng, H.; Xu, J.; Wang, J.; Yuan, M.S. Employing a fluorescent and colorimetric picolyl-functionalized rhodamine for the detection of glyphosate pesticide. Talanta 2021, 224, 121834. [Google Scholar] [CrossRef]
- Sasaki, Y.; Asano, K.; Minamiki, T.; Zhang, Z.; Takizawa, S.Y.; Kubota, R.; Minami, T. A Water-Gated Organic Thin-Film Transistor for Glyphosate Detection: A Comparative Study with Fluorescence Sensing. Chemistry 2020, 26, 14506. [Google Scholar] [CrossRef]
- Habekost, A. Rapid and sensitive spectroelectrochemical and electrochemical detection of glyphosate and AMPA with screen-printed electrodes. Talanta 2017, 162, 583–588. [Google Scholar] [CrossRef]
- Sahoo, D.; Mandal, A.; Mitra, T.; Chakraborty, K.; Bardhan, M.; Dasgupta, A.K. Nanosensing of Pesticides by Zinc Oxide Quantum Dot: An Optical and Electrochemical Approach for the Detection of Pesticides in Water. J. Agric. Food Chem. 2018, 66, 414–423. [Google Scholar] [CrossRef]
- Kalra, P.; Dhiman, A.; Cho, W.C.; Bruno, J.G.; Sharma, T.K. Simple Methods and Rational Design for Enhancing Aptamer Sensitivity and Specificity. Front. Mol. Biosci. 2018, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Shaban, S.M.; Kim, D.H. Recent Advances in Aptamer Sensors. Sensors 2021, 21, 979. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.K.; Bruno, J.G.; Dhiman, A. ABCs of DNA aptamer and related assay development. Biotechnol. Adv. 2017, 35, 275–301. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jiang, Y.; Cui, C.; Yang, Y.; Zhang, P.; Stewart, K.; Pan, X.; Li, X.; Yang, L.; Qiu, L.; et al. Modulating Aptamer Specificity with pH-Responsive DNA Bonds. J. Am. Chem. Soc. 2018, 140, 13335–13339. [Google Scholar] [CrossRef]
- Ojha, Y.R.; Giovannucci, D.R.; Cameron, B.D. Selection and characterization of structure-switching DNA aptamers for the salivary peptide histatin 3. J. Biotechnol. 2021, 327, 9–17. [Google Scholar] [CrossRef]
- Ren, Q.; Mou, J.; Guo, Y.; Wang, H.; Cao, X.; Zhang, F.; Xia, J.; Wang, Z. Simple homogeneous electrochemical target-responsive aptasensor based on aptamer bio-gated and porous carbon nanocontainer derived from ZIF-8. Biosens. Bioelectron. 2020, 166, 112448. [Google Scholar] [CrossRef]
- Dong, H.; Chen, H.; Jiang, J.; Zhang, H.; Cai, C.; Shen, Q. Highly Sensitive Electrochemical Detection of Tumor Exosomes Based on Aptamer Recognition-Induced Multi-DNA Release and Cyclic Enzymatic Amplification. Anal. Chem. 2018, 90, 4507–4513. [Google Scholar] [CrossRef]
- Shi, H.; He, X.; Wang, K.; Wu, X.; Ye, X.; Guo, Q.; Tan, W.; Qing, Z.; Yang, X.; Zhou, B. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc. Natl. Acad. Sci. USA 2011, 108, 3900–3905. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.J.; Cheng, F.F.; Zheng, T.T.; Zhu, J.J. Versatile aptasensor for electrochemical quantification of cell surface glycan and naked-eye tracking glycolytic inhibition in living cells. Biosens. Bioelectron. 2017, 89 Pt 2, 937–945. [Google Scholar] [CrossRef]
- Selvolini, G.; Bajan, I.; Hosu, O.; Cristea, C.; Sandulescu, R.; Marrazza, G. DNA-Based Sensor for the Detection of an Organophosphorus Pesticide: Profenofos. Sensors 2018, 18, 2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, C.; Zhang, X.; Ye, S.; Yang, H.; Huang, Z.; Yang, D.; Cai, R.; Tan, W. Aptamer-Pendant DNA Tetrahedron Nanostructure Probe for Ultrasensitive Detection of Tetracycline by Coupling Target-Triggered Rolling Circle Amplification. ACS Appl. Mater. Interfaces 2021, 13, 19695–19700. [Google Scholar] [CrossRef] [PubMed]
- Trinh, K.H.; Kadam, U.S.; Rampogu, S.; Cho, Y.; Yang, K.A.; Kang, C.H.; Lee, K.W.; Lee, K.O.; Chung, W.S.; Hong, J.C. Development of novel fluorescence-based and label-free noncanonical G4-quadruplex-like DNA biosensor for facile, specific, and ultrasensitive detection of fipronil. J. Hazard Mater. 2022, 427, 127939. [Google Scholar] [CrossRef] [PubMed]
- Trinh, K.H.; Kadam, U.S.; Song, J.; Cho, Y.; Kang, C.H.; Lee, K.O.; Lim, C.O.; Chung, W.S.; Hong, J.C. Novel DNA Aptameric Sensors to Detect the Toxic Insecticide Fenitrothion. Int. J. Mol. Sci. 2021, 22, 10846. [Google Scholar] [CrossRef]
- Saito, T.; Aoki, H.; Namera, A.; Oikawa, H.; Miyazaki, S.; Nakamoto, A.; Inokuchi, S. Mix-mode TiO-C18 monolith spin column extraction and GC-MS for the simultaneous assay of organophosphorus compounds and glufosinate, and glyphosate in human serum and urine. Anal. Sci. 2011, 27, 999–1005. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Lin, B.; Cao, Y.; Guo, M.; Yu, Y. A Highly Selective and Sensitive Fluorescence Detection Method of Glyphosate Based on an Immune Reaction Strategy of Carbon Dot Labeled Antibody and Antigen Magnetic Beads. J. Agric. Food Chem. 2016, 64, 6042–6050. [Google Scholar] [CrossRef]
- Vu, C.T.; Le, P.T.; Chu, D.B.; Bui, V.H.; Phung, T.L.A.; Nguyen Le, H.Y.; Labanowski, J.; Mondamert, L.; Herrmann, M.; Behra, P. One-step purification/extraction method to access glyphosate, glufosinate, and their metabolites in natural waters. J. Chromatogr. A 2021, 1649, 462188. [Google Scholar] [CrossRef]
- Druart, C.; Delhomme, O.; de Vaufleury, A.; Ntcho, E.; Millet, M. Optimization of extraction procedure and chromatographic separation of glyphosate, glufosinate and aminomethylphosphonic acid in soil. Anal. Bioanal. Chem. 2011, 399, 1725–1732. [Google Scholar] [CrossRef]
- Pareja, L.; Jesús, F.; Heinzen, H.; Hernando, M.D.; Rajski, Ł.; Fernández-Alba, A.R. Evaluation of glyphosate and AMPA in honey by water extraction followed by ion chromatography mass spectrometry. A pilot monitoring study. Anal. Methods 2019, 11, 2123–2128. [Google Scholar] [CrossRef]
- Lanaro, R.; Costa, J.L.; Cazenave, S.O.; Zanolli-Filho, L.A.; Tavares, M.F.; Chasin, A.A. Determination of herbicides paraquat, glyphosate, and aminomethylphosphonic acid in marijuana samples by capillary electrophoresis. J. Forensic. Sci. 2015, 60 (Suppl. 1), S241–S247. [Google Scholar] [CrossRef]
- Steinborn, A.; Alder, L.; Michalski, B.; Zomer, P.; Bendig, P.; Martinez, S.A.; Mol, H.G.; Class, T.J.; Pinheiro, N.C. Determination of Glyphosate Levels in Breast Milk Samples from Germany by LC-MS/MS and GC-MS/MS. J. Agric. Food Chem. 2016, 64, 1414–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; She, Y.; Li, T.; Zhao, F.; Jin, M.; Guo, Y.; Zheng, L.; Wang, S.; Jin, F.; Shao, H.; et al. A highly selective electrochemical sensor based on molecularly imprinted polypyrrole-modified gold electrode for the determination of glyphosate in cucumber and tap water. Anal. Bioanal. Chem. 2017, 409, 7133–7144. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, G.; Liu, H.; Leung, C.-H.; Ma, D.-L. G-quadruplex-based detection of glyphosate in complex biological systems by a time-resolved luminescent assay. Sens. Actuators B Chem. 2020, 320, 128393. [Google Scholar] [CrossRef]

| Method | LOD | Recovery | Sample | Reference |
|---|---|---|---|---|
| GC–MS | 3 μmol/L | 96.7–107.7% | Serum and urine | [45] |
| ELISA | 0.047μmol/L | 87.4–103.7% | River water, Tea and soil | [46] |
| HPLC | 0.004 μmol/L | 80.1–109.4% | Natural water | [47] |
| LC | 6 μmol/L | 80.63–97.11% | Soil | [48] |
| IC | 30 μmol/L | 80–110% | Honey | [49] |
| CE | 800 mmol/L | 89.4–93.7% | Hemp | [50] |
| LC–MS/MS | 6 μmol/L | 97–110% | Breast milk | [51] |
| EC | 0.16 mmol/L | 72.7–98.96% | Cucumber Tap water | [52] |
| qPCR | 0.6 μmol/L | 103.4–104.9% | Tap water | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, Y.; Tian, R.; Duan, J.; Wang, M.; Cao, J.; Cao, Z.; Li, G.; Jin, F.; Abd El-Aty, A.M.; She, Y. A Novel Fluorescent Sensor Based on Aptamer and qPCR for Determination of Glyphosate in Tap Water. Sensors 2023, 23, 649. https://doi.org/10.3390/s23020649
Shao Y, Tian R, Duan J, Wang M, Cao J, Cao Z, Li G, Jin F, Abd El-Aty AM, She Y. A Novel Fluorescent Sensor Based on Aptamer and qPCR for Determination of Glyphosate in Tap Water. Sensors. 2023; 23(2):649. https://doi.org/10.3390/s23020649
Chicago/Turabian StyleShao, Yong, Run Tian, Jiaqi Duan, Miao Wang, Jing Cao, Zhen Cao, Guangyue Li, Fen Jin, A. M. Abd El-Aty, and Yongxin She. 2023. "A Novel Fluorescent Sensor Based on Aptamer and qPCR for Determination of Glyphosate in Tap Water" Sensors 23, no. 2: 649. https://doi.org/10.3390/s23020649






