A Comprehensive Landscape of Imaging Feature-Associated RNA Expression Profiles in Human Breast Tissue
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Image Data Processiong
2.3. Unsupervised Analysis of Breast Images Using Nucleus Features
2.4. Identification of Feature-Specific Genes
3. Results
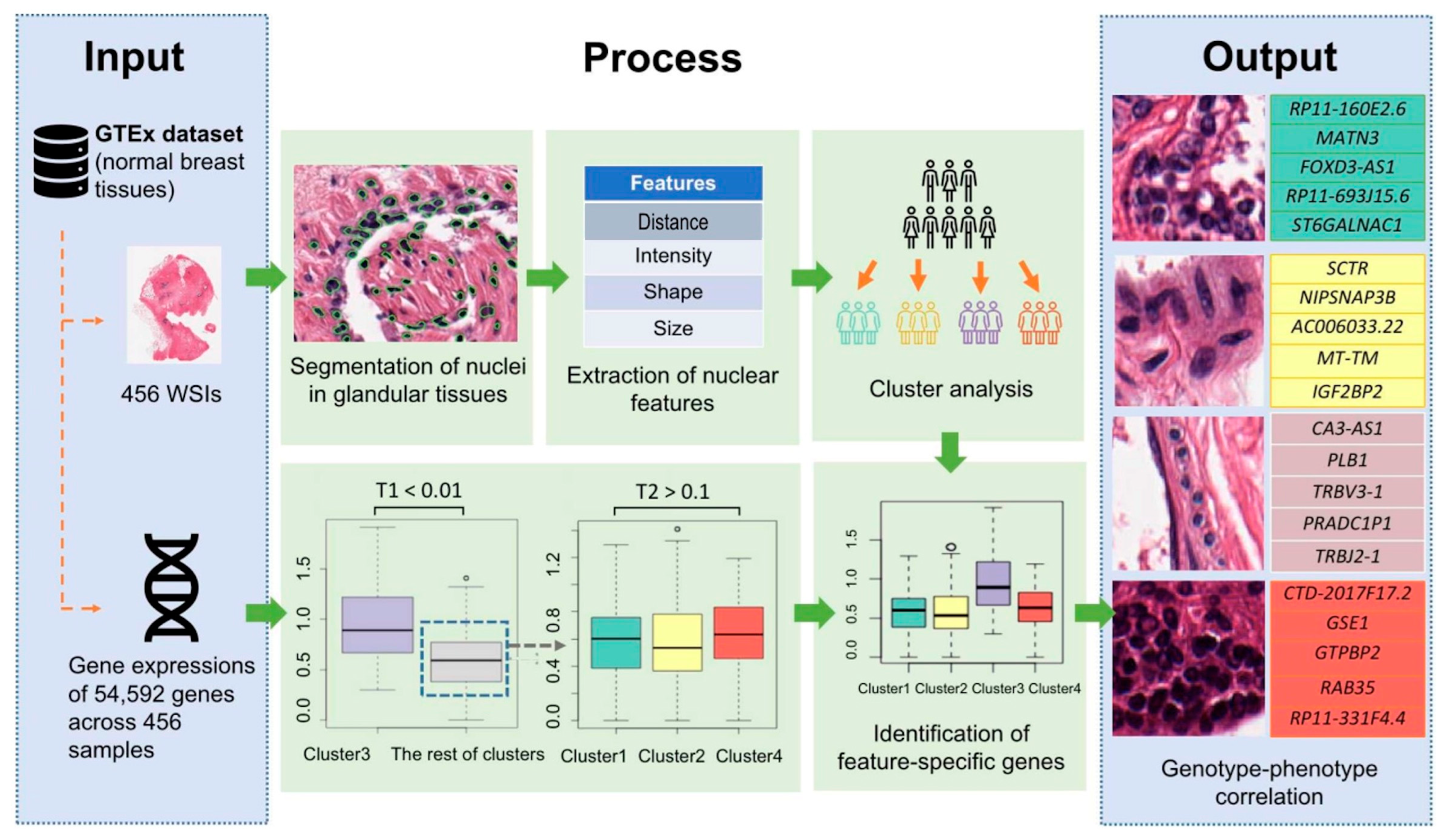
3.1. Image-Genetic Joint Analysis Pipeline
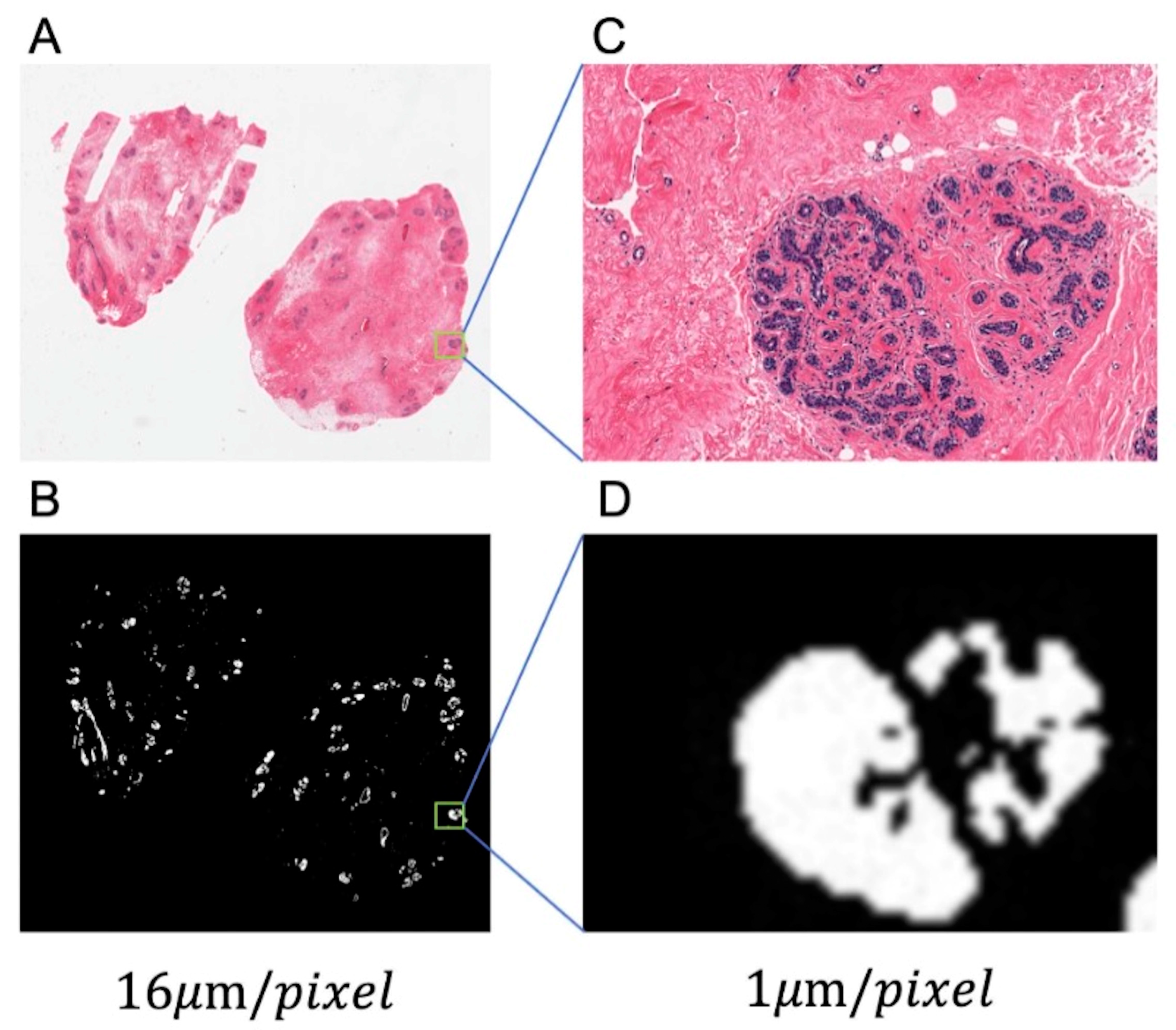
3.2. Glandular Tissue Segmentation
3.3. Nuclei Segmentation
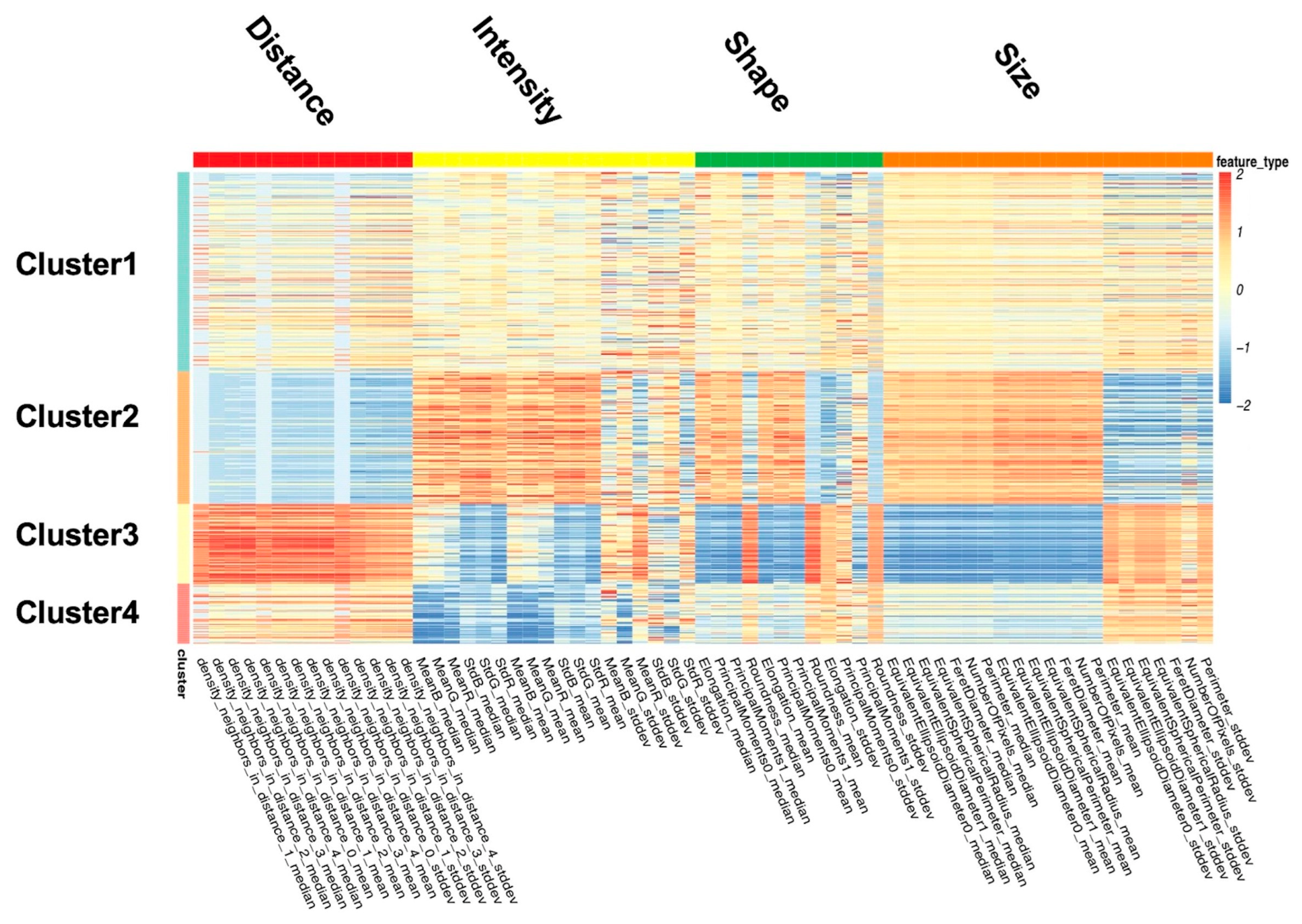
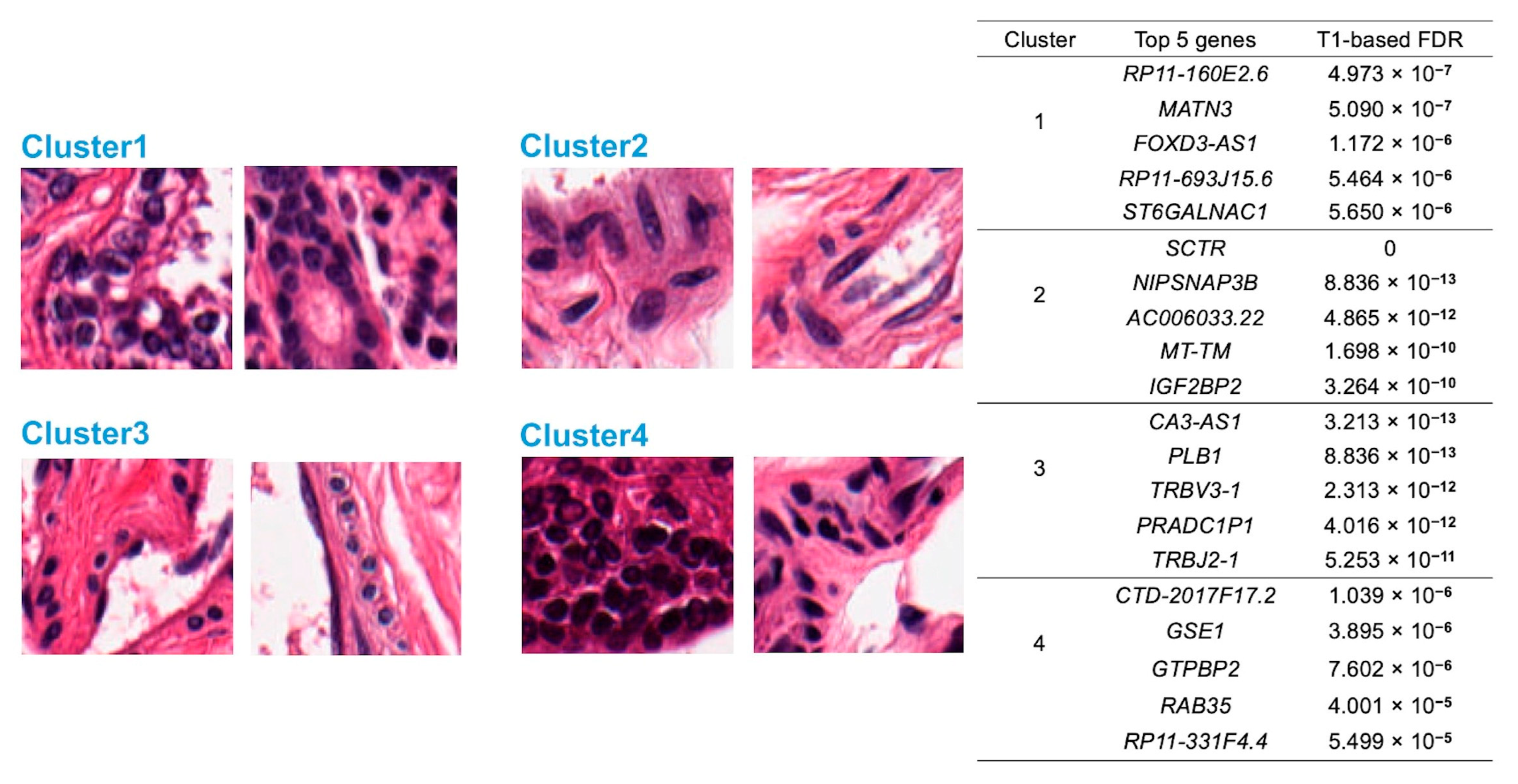
3.4. Classification of Image Features
- Cluster 1: All nuclear features are close to the sample mean.
- Cluster 2: The nuclei in this cluster are large, irregular, long, and dark, with the most uneven color distribution. The distance between the nuclei is small.
- Cluster 3: The nuclei in this cluster are small and round, with uniform color distribution. The distances among the nuclei are large.
- Cluster 4: The nuclei in this cluster appear to be quite dark.
3.5. Discovery of Feature-Specific Genes
3.6. Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhan, X.; Cheng, J.; Huang, Z.; Han, Z.; Helm, B.; Liu, X.; Zhang, J.; Wang, T.-F.; Ni, D.; Huang, K. Correlation Analysis of Histopathology and Proteogenomics Data for Breast Cancer. Mol. Cell. Proteom. 2019, 18, S37–S51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, L.K.; Cristofanilli, M.; Zhou, X.; Welch, K.L.; Smith, T.L.; Yang, Y.; Sneige, N.; Sahin, A.A.; Gilcrease, M.Z. β4 integrin subunit gene expression correlates with tumor size and nuclear grade in early breast cancer. Mod. Pathol. 2005, 18, 1165–1175. [Google Scholar] [CrossRef] [Green Version]
- Bianchini, G.; Qi, Y.; Alvarez, R.H.; Iwamoto, T.; Coutant, C.; Ibrahim, N.K.; Valero, V.; Cristofanilli, M.; Green, M.C.; Radvanyi, L.; et al. Molecular Anatomy of Breast Cancer Stroma and Its Prognostic Value in Estrogen Receptor–Positive and –Negative Cancers. J. Clin. Oncol. 2010, 28, 4316–4323. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2010, 5, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.H.; Sangoi, A.R.; Leung, S.; Marinelli, R.J.; Nielsen, T.O.; van de Vijver, M.J.; West, R.B.; van de Rijn, M.; Koller, D. Systematic Analysis of Breast Cancer Morphology Uncovers Stromal Features Associated with Survival. Sci. Transl. Med. 2011, 3, 108ra113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ash, J.T.; Darnell, G.; Munro, D.; Engelhardt, B.E. Joint analysis of expression levels and histological images identifies genes associated with tissue morphology. Nat. Commun. 2021, 12, 1609. [Google Scholar] [CrossRef] [PubMed]
- Irshad, H.; Veillard, A.; Roux, L.; Racoceanu, D. Methods for Nuclei Detection, Segmentation, and Classification in Digital Histopathology: A Review—Current Status and Future Potential. IEEE Rev. Biomed. Eng. 2013, 7, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Veta, M.M.; Van Diest, P.J.; Kornegoor, R.; Huisman, A.; Viergever, M.A.; Pluim, J.P.W. Automatic Nuclei Segmentation in H&E Stained Breast Cancer Histopathology Images. PLoS ONE 2013, 8, e70221. [Google Scholar] [CrossRef] [Green Version]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [Green Version]
- Sirinukunwattana, K.; Raza, S.E.A.; Tsang, Y.-W.; Snead, D.R.J.; Cree, I.A.; Rajpoot, N.M. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206. [Google Scholar] [CrossRef]
- Wang, P.; Hu, X.; Li, Y.; Liu, Q.; Zhu, X. Automatic cell nuclei segmentation and classification of breast cancer histopathology images. Signal Process. 2016, 122, 1–13. [Google Scholar] [CrossRef]
- Sun, D.; Li, A.; Tang, B.; Wang, M. Integrating genomic data and pathological images to effectively predict breast cancer clinical outcome. Comput. Methods Programs Biomed. 2018, 161, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, H.; Zhou, X. Nuclei Segmentation Using Marker-Controlled Watershed, Tracking Using Mean-Shift, and Kalman Filter in Time-Lapse Microscopy. IEEE Trans. Circuits Syst. I: Regul. Pap. 2006, 53, 2405–2414. [Google Scholar] [CrossRef]
- Naik, S.; Doyle, S.; Feldman, M.; Tomaszewski, J.; Madabhushi, A. Gland segmentation and computerized gleason grading of prostate histology by integrating low-, high-level and domain specific information. In MIAAB Workshop; Citeseer: Piscataway, NJ, USA, 2007; pp. 1–8. [Google Scholar]
- Isaksson, J.; Arvidsson, I.; Aastrom, K.; Heyden, A. Semantic Segmentation of Microscopic Images of H&E Stained Prostatic Tissue Using CNN; IEEE: Toulouse, France, 2017; pp. 1252–1256. [Google Scholar] [CrossRef]
- Johnson, J.W. Adapting mask-rcnn for automatic nucleus segmentation. arXiv preprint 2018, arXiv:1805.00500. [Google Scholar] [CrossRef]
- Vuola, A.O.; Akram, S.U.; Kannala, J. Mask-RCNN and U-net ensembled for nuclei segmentation. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 208–212. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics international 2004, 11, 36–42. [Google Scholar]
- Bahlmann, C.; Patel, A.; Johnson, J.; Ni, J.; Chekkoury, A.; Khurd, P.; Kamen, A.; Grady, L.; Krupinski, E.; Graham, A.; et al. Automated detection of diagnostically relevant regions in H&E stained digital pathology slides. Prog. Biomed. Opt. Imaging 2012, 8315, 831504. [Google Scholar] [CrossRef]
- Linder, N.; Konsti, J.; Turkki, R.; Rahtu, E.; Lundin, M.; Nordling, S.; Haglund, C.; Ahonen, T.; Pietikäinen, M.; Lundin, J. Identification of tumor epithelium and stroma in tissue microarrays using texture analysis. Diagn. Pathol. 2012, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- McKenna, S.J.; Amaral, T.; Akbar, S.; Jordan, L.; Thompson, A. Immunohistochemical analysis of breast tissue microarray images using contextual classifiers. J. Pathol. Informatics 2013, 4, 13. [Google Scholar] [CrossRef]
- Vink, J.P.; Van Leeuwen, M.B.; Van Deurzen, C.H.M.; De Haan, G.G. Efficient nucleus detector in histopathology images. J. Microsc. 2013, 249, 124–135. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.M.; El-Daly, H.; Simmons, E.; Rajpoot, N.M. HyMaP: A hybrid magnitude-phase approach to unsupervised segmentation of tumor areas in breast cancer histology images. J. Pathol. Informatics 2013, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Kim, C.; Chae, S.W.; Oh, S. Unsupervised Segmentation of Overlapped Nuclei Using Bayesian Classification. IEEE Trans. Biomed. Eng. 2010, 57, 2825–2832. [Google Scholar] [CrossRef] [PubMed]
- Saltz, J.; Almeida, J.; Gao, Y.; Sharma, A.; Bremer, E.; DiPrima, T.; Saltz, M.; Kalpathy-Cramer, J.; Kurc, T. Towards Generation, Management, and Exploration of Combined Radiomics and Pathomics Datasets for Cancer Research. AMIA Jt. Summits Transl. Sci. proceedings. AMIA Jt. Summits Transl. Sci. 2017, 2017, 85–94. [Google Scholar]
- Wen, S.; Kurc, T.M.; Gao, Y.; Zhao, T.; Saltz, J.H.; Zhu, W. A Methodology for Texture Feature-based Quality Assessment in Nucleus Segmentation of Histopathology Image. J. Pathol. Informatics 2017, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Han, Z.; Mehra, R.; Shao, W.; Cheng, M.; Feng, Q.; Ni, D.; Huang, K.; Cheng, L.; Zhang, J. Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11.2 translocation renal cell carcinoma. Nat. Commun. 2020, 11, 1778. [Google Scholar] [CrossRef] [Green Version]
- Blaveri, E.; Simko, J.P.; Korkola, J.E.; Brewer, J.L.; Baehner, F.; Mehta, K.; DeVries, S.; Koppie, T.; Pejavar, S.; Carroll, P.; et al. Bladder Cancer Outcome and Subtype Classification by Gene Expression. Clin. Cancer Res. 2005, 11, 4044–4055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sotiriou, C.; Pusztai, L. Gene-Expression Signatures in Breast Cancer. New Engl. J. Med. 2009, 360, 790–800. [Google Scholar] [CrossRef] [Green Version]
- Barry, W.T.; Kernagis, D.N.; Dressman, H.K.; Griffis, R.J.; Hunter, J.D.; Olson, J.A.; Marks, J.R.; Ginsburg, G.S.; Marcom, P.K.; Nevins, J.R.; et al. Intratumor Heterogeneity and Precision of Microarray-Based Predictors of Breast Cancer Biology and Clinical Outcome. J. Clin. Oncol. 2010, 28, 2198–2206. [Google Scholar] [CrossRef] [Green Version]
- Gevaert, O.; Xu, J.; Hoang, C.D.; Leung, A.N.; Xu, Y.; Quon, A.; Rubin, D.L.; Napel, S.; Plevritis, S.K. Non–Small Cell Lung Cancer: Identifying Prognostic Imaging Biomarkers by Leveraging Public Gene Expression Microarray Data—Methods and Preliminary Results. Radiology 2012, 264, 387–396. [Google Scholar] [CrossRef]
- Chen, R.J.; Lu, M.Y.; Wang, J.; Williamson, D.F.K.; Rodig, S.J.; Lindeman, N.I.; Mahmood, F. Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and Prognosis. IEEE Trans. Med. Imaging 2022, 41, 757–770. [Google Scholar] [CrossRef]
- Dolezal, J.M.; Trzcinska, A.; Liao, C.-Y.; Kochanny, S.; Blair, E.; Agrawal, N.; Keutgen, X.M.; Angelos, P.; Cipriani, N.A.; Pearson, A.T. Deep learning prediction of BRAF-RAS gene expression signature identifies noninvasive follicular thyroid neoplasms with papillary-like nuclear features. Mod. Pathol. 2021, 34, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Cirera, L.; Cabana-Domínguez, J.; Lee, P.H.; Fernàndez-Castillo, N.; Cormand, B. Identification of genetic variants influencing methylation in brain with pleiotropic effects on psychiatric disorders. Prog. Neuro-Psychopharmacology Biol. Psychiatry 2021, 113, 110454. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, P.J. Robust statistics. In International encyclopedia of statistical science; Springer: Berlin, Heidelberg, 2011; pp. 1248–1251. [Google Scholar]
- Pichon, E.; Tannenbaum, A.; Kikinis, R. A statistically based flow for image segmentation. Med. Image Anal. 2004, 8, 267–274. [Google Scholar] [CrossRef]
- Gao, Y.; Kikinis, R.; Bouix, S.; Shenton, M.; Tannenbaum, A. A 3D interactive multi-object segmentation tool using local robust statistics driven active contours. Med. Image Anal. 2012, 16, 1216–1227. [Google Scholar] [CrossRef] [Green Version]
- Ruifrok, A.C.; Johnston, D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001, 23, 291–299. [Google Scholar]
- Vese, L.A.; Chan, T.F. A Multiphase Level Set Framework for Image Segmentation Using the Mumford and Shah Model. Int. J. Comput. Vis. 2002, 50, 271–293. [Google Scholar] [CrossRef]
- Qi, X.; Xing, F.; Foran, D.J.; Yang, L. Robust Segmentation of Overlapping Cells in Histopathology Specimens Using Parallel Seed Detection and Repulsive Level Set. IEEE Trans. Biomed. Eng. 2011, 59, 754–765. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, G. Label object representation and manipulation with ITK. Insight J. 2007. [Google Scholar] [CrossRef]
- Doyle, S.; Hwang, M.; Shah, K.; Madabhushi, A.; Feldman, M.; Tomaszeweski, J. Automated Grading Of Prostate Cancer Using Architectural And Textural Image Features; IEEE: Toulouse, France, 2007; pp. 1284–1287. [Google Scholar] [CrossRef]
- Caron, M.; Bojanowski, P.; Joulin, A.; Douze, M. Deep clustering for unsupervised learning of visual features. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 132–149. [Google Scholar] [CrossRef]
- Min, E.; Guo, X.; Liu, Q.; Zhang, G.; Cui, J.; Long, J. A Survey of Clustering With Deep Learning: From the Perspective of Network Architecture. IEEE Access 2018, 6, 39501–39514. [Google Scholar] [CrossRef]
- MacQueen, J. Classification and analysis of multivariate observations. 5th Berkeley Symp. Math. Statist. Probability 1967, 281–297. [Google Scholar]
- Vu, T.N.; Pramana, S.; Calza, S.; Suo, C.; Lee, D.; Pawitan, Y. Comprehensive landscape of subtype-specific coding and non-coding RNA transcripts in breast cancer. Oncotarget 2016, 7, 68851–68863. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Pawitan, Y.; Michiels, S.; Koscielny, S.; Gusnanto, A.; Ploner, A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics 2005, 21, 3017–3024. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef] [Green Version]
- Medical Image Computing C.A.I.S., 2016. Miccai2016. Available online: http://www.miccai2016.org/en (accessed on 13 October 2021).
- Sirinukunwattana, K.; Pluim, J.P.; Chen, H.; Qi, X.; Heng, P.-A.; Guo, Y.B.; Wang, L.Y.; Matuszewski, B.J.; Bruni, E.; Sanchez, U.; et al. Gland segmentation in colon histology images: The glas challenge contest. Med. Image Anal. 2017, 35, 489–502. [Google Scholar] [CrossRef] [Green Version]
- Maška, M.; Ulman, V.; Svoboda, D.; Matula, P.; Matula, P.; Ederra, C.; Urbiola, A.; España, T.; Venkatesan, S.; Balak, D.M.; et al. A benchmark for comparison of cell tracking algorithms. Bioinformatics 2014, 30, 1609–1617. [Google Scholar] [CrossRef] [Green Version]
- Caicedo, J.C.; Goodman, A.; Karhohs, K.W.; Cimini, B.A.; Ackerman, J.; Haghighi, M.; Heng, C.; Becker, T.; Doan, M.; McQuin, C.; et al. Nucleus segmentation across imaging experiments: The 2018 Data Science Bowl. Nat. Methods 2019, 16, 1247–1253. [Google Scholar] [CrossRef] [Green Version]
- Mou, T.; Pawitan, Y.; Stahl, M.; Vesterlund, M.; Deng, W.; Jafari, R.; Bohlin, A.; Österroos, A.; Siavelis, L.; Bäckvall, H.; et al. The transcriptome-wide landscape of molecular subtype-specific mRNA expression profiles in acute myeloid leukemia. Am. J. Hematol. 2021, 96, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Onori, P.; Wise, C.; Gaudio, E.; Franchitto, A.; Francis, H.; Carpino, G.; Lee, V.; Lam, I.; Miller, T.; Dostal, D.E.; et al. Secretin inhibits cholangiocarcinoma growth via dysregulation of the cAMP-dependent signaling mechanisms of secretin receptor. Int. J. Cancer 2009, 127, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, L.; Fu, J.; Huang, H.; Sun, S.; Zhang, D.; Zhao, L.; Onwuka, J.U.; Zhao, Y.; Cui, B. SCTR hypermethylation is a diagnostic biomarker in colorectal cancer. Cancer Sci. 2020, 111, 4558–4566. [Google Scholar] [CrossRef]
- Kang, S.; Kim, B.; Kang, H.-S.; Jeong, G.; Bae, H.; Lee, H.; Lee, S.; Kim, S.J. SCTR regulates cell cycle-related genes toward anti-proliferation in normal breast cells while having pro-proliferation activity in breast cancer cells. Int. J. Oncol. 2015, 47, 1923–1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMullen, E.R.; Gonzalez, M.E.; Skala, S.L.; Tran, M.; Thomas, D.; Djomehri, S.I.; Burman, B.; Kidwell, K.M.; Kleer, C.G. CCN6 regulates IGF2BP2 and HMGA2 signaling in metaplastic carcinomas of the breast. Breast Cancer Res. Treat. 2018, 172, 577–586. [Google Scholar] [CrossRef]
- Kim, H.-Y.; Thi, H.T.H.; Hong, S. IMP2 and IMP3 cooperate to promote the metastasis of triple-negative breast cancer through destabilization of progesterone receptor. Cancer Lett. 2018, 415, 30–39. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Zhuang, H.-W.; Wang, J.; Shen, Y.; Bu, Y.-Z.; Guan, B.-G.; Xu, F.; Dou, J. Long noncoding RNA CA3-AS1 suppresses gastric cancer migration and invasion by sponging miR-93-5p and targeting BTG3. Gene Ther. 2020, 29, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yang, Z.; Lin, B. Overexpression of long non coding RNA CA3-AS1 suppresses proliferation, invasion and promotes apoptosis via miRNA-93/PTEN axis in colorectal cancer. Gene 2018, 687, 9–15. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [Green Version]
- Hajizadeh, F.; Maleki, L.A.; Alexander, M.; Mikhailova, M.V.; Masjedi, A.; Ahmadpour, M.; Hashemi, V.; Jadidi-Niaragh, F. Tumor-associated neutrophils as new players in immunosuppressive process of the tumor microenvironment in breast cancer. Life Sci. 2020, 264, 118699. [Google Scholar] [CrossRef]
- Mollinedo, F. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol. 2019, 40, 228–242. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Psyrri, A.; Kalogeras, K.; Kronenwett, R.; Wirtz, R.; Batistatou, A.; Bournakis, E.; Timotheadou, E.; Gogas, H.; Aravantinos, G.; Christodoulou, C.; et al. Prognostic significance of UBE2C mRNA expression in high-risk early breast cancer. A Hellenic Cooperative Oncology Group (HeCOG) Study. Ann. Oncol. 2012, 23, 1422–1427. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Zhou, C.; Liu, F.; Xu, G.; Zheng, R.; Zhang, X. MicroRNA-196a post-transcriptionally upregulates the UBE2C proto-oncogene and promotes cell proliferation in breast cancer. Oncol. Rep. 2015, 34, 877–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bièche, I.; Vacher, S.; Lallemand, F.; Tozlu-Kara, S.; Bennani, H.; Beuzelin, M.; Driouch, K.; Rouleau, E.; Lerebours, F.; Ripoche, H.; et al. Expression analysis of mitotic spindle checkpoint genes in breast carcinoma: Role of NDC80/HEC1 in early breast tumorigenicity, and a two-gene signature for aneuploidy. Mol. Cancer 2011, 10, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Wang, Y.; Wang, Y.; Lv, S.; Xu, X.; Dong, X. Screening of differentially expressed genes and identification of NUF2 as a prognostic marker in breast cancer. Int. J. Mol. Med. 2019, 44, 390–404. [Google Scholar] [CrossRef] [Green Version]
- Opoku, F.; Bedu-Addo, K.; Titiloye, N.A.; Manu, E.A.; Ameh-Mensah, C.; Duduyemi, B.M. Expression profile of tumour suppressor protein p53 and its regulator MDM2 in a cohort of breast cancer patients in a Tertiary Hospital in Ghana. PLoS ONE 2021, 16, e0258543. [Google Scholar] [CrossRef]
- Wege, A.K.; Rom-Jurek, E.; Jank, P.; Denkert, C.; Ugocsai, P.; Solbach, C.; Blohmer, J.; Sinn, B.; Mackelenbergh, M.; Möbus, V.; et al. mdm2 gene amplification is associated with luminal breast cancer progression in humanized PDX mice and a worse outcome of estrogen receptor positive disease. Int. J. Cancer 2021, 150, 1357–1372. [Google Scholar] [CrossRef]
- Ethier, J.-L.; Desautels, D.; Templeton, A.; Shah, P.S.; Amir, E. Prognostic role of neutrophil-to-lymphocyte ratio in breast cancer: A systematic review and meta-analysis. Breast Cancer Res. 2017, 19, 2. [Google Scholar] [CrossRef] [Green Version]
- Kakinuma, T. Chemokines, chemokine receptors, and cancer metastasis. J. Leukoc. Biol. 2006, 79, 639–651. [Google Scholar] [CrossRef]
- Pour, A.F.; White, B.S.; Park, J.; Sheridan, T.B.; Chuang, J.H. Deep learning features encode interpretable morphologies within histological images. Sci. Rep. 2022, 12, 9428. [Google Scholar] [CrossRef]





| DSC (mean ± std) | IoU (mean ± std) | Average HD (mean ± std) | |
|---|---|---|---|
| QuPath | 0.7002 ± 0.0904 | 0.5451 ± 0.1006 | 2.8108 ± 2.0811 |
| UNet | 0.7592 ± 0.0983 | 0.6204 ± 0.1187 | 1.3044 ± 0.7208 |
| Proposed | 0.7797 ± 0.0525 | 0.6416 ± 0.0691 | 1.2942 ± 0.6634 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mou, T.; Liang, J.; Vu, T.N.; Tian, M.; Gao, Y. A Comprehensive Landscape of Imaging Feature-Associated RNA Expression Profiles in Human Breast Tissue. Sensors 2023, 23, 1432. https://doi.org/10.3390/s23031432
Mou T, Liang J, Vu TN, Tian M, Gao Y. A Comprehensive Landscape of Imaging Feature-Associated RNA Expression Profiles in Human Breast Tissue. Sensors. 2023; 23(3):1432. https://doi.org/10.3390/s23031432
Chicago/Turabian StyleMou, Tian, Jianwen Liang, Trung Nghia Vu, Mu Tian, and Yi Gao. 2023. "A Comprehensive Landscape of Imaging Feature-Associated RNA Expression Profiles in Human Breast Tissue" Sensors 23, no. 3: 1432. https://doi.org/10.3390/s23031432





