Biosensing with Luminescent Semiconductor Quantum Dots
Abstract
:1. Introduction
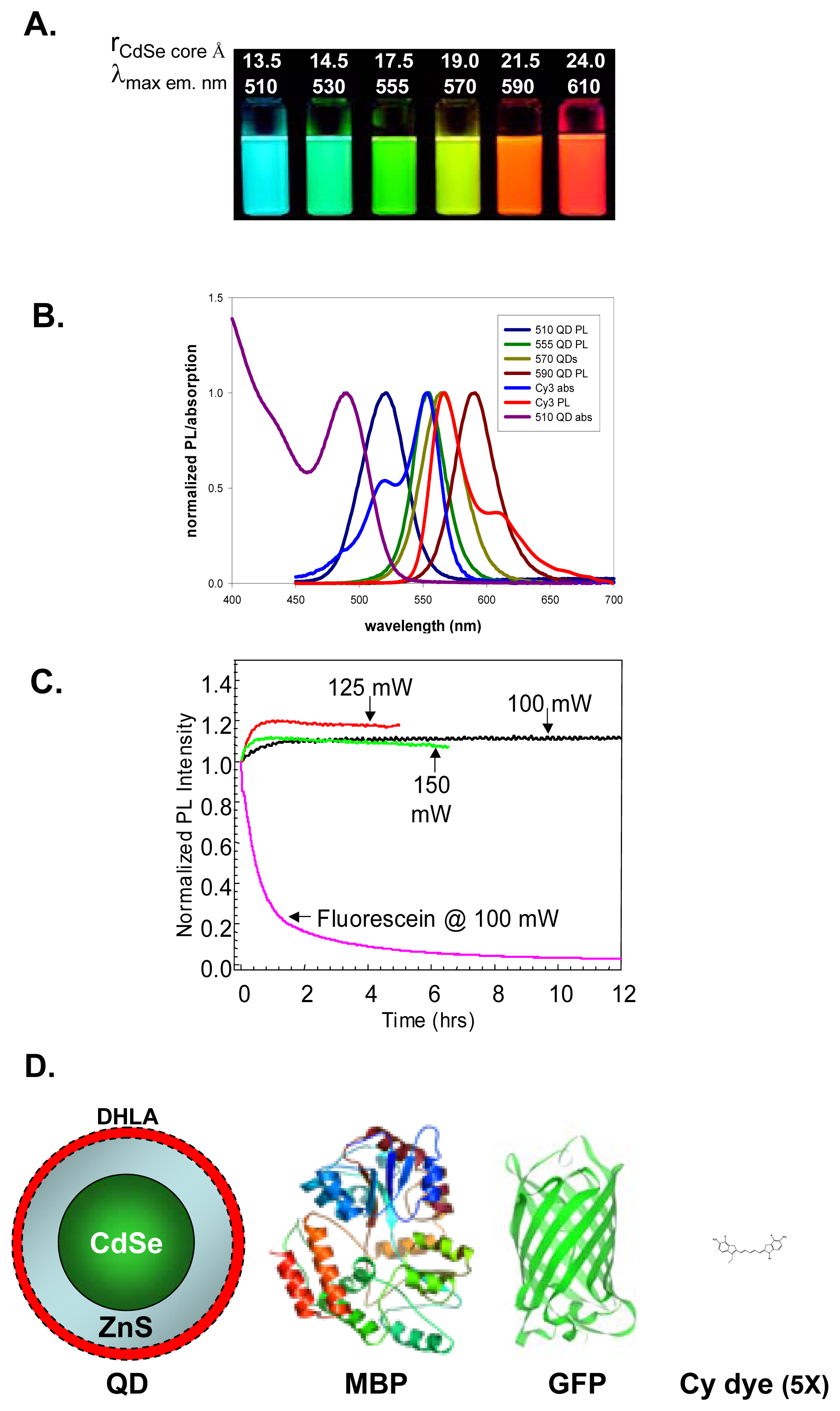
2. Synthesis and Properties of Luminescent Quantum Dots
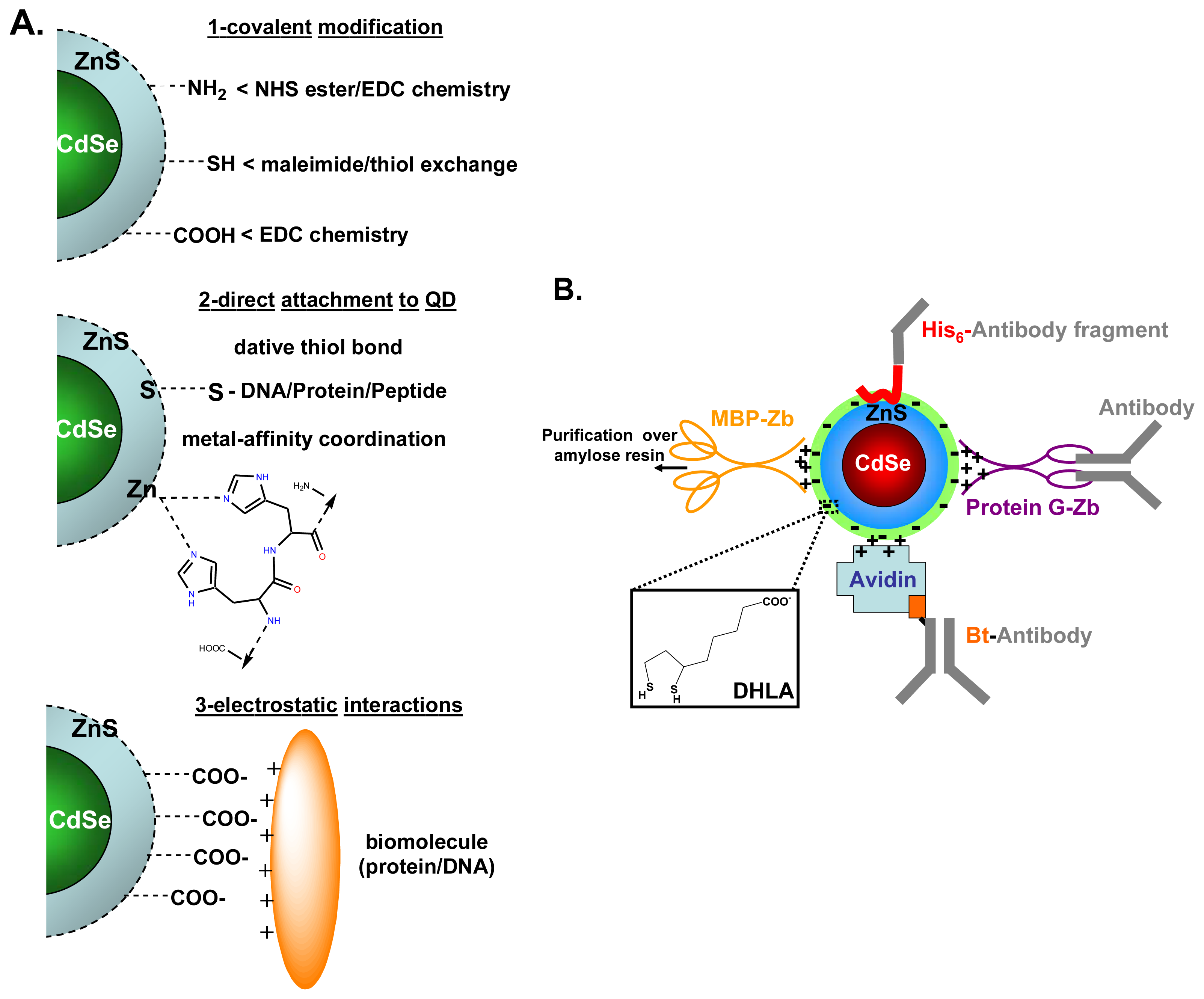
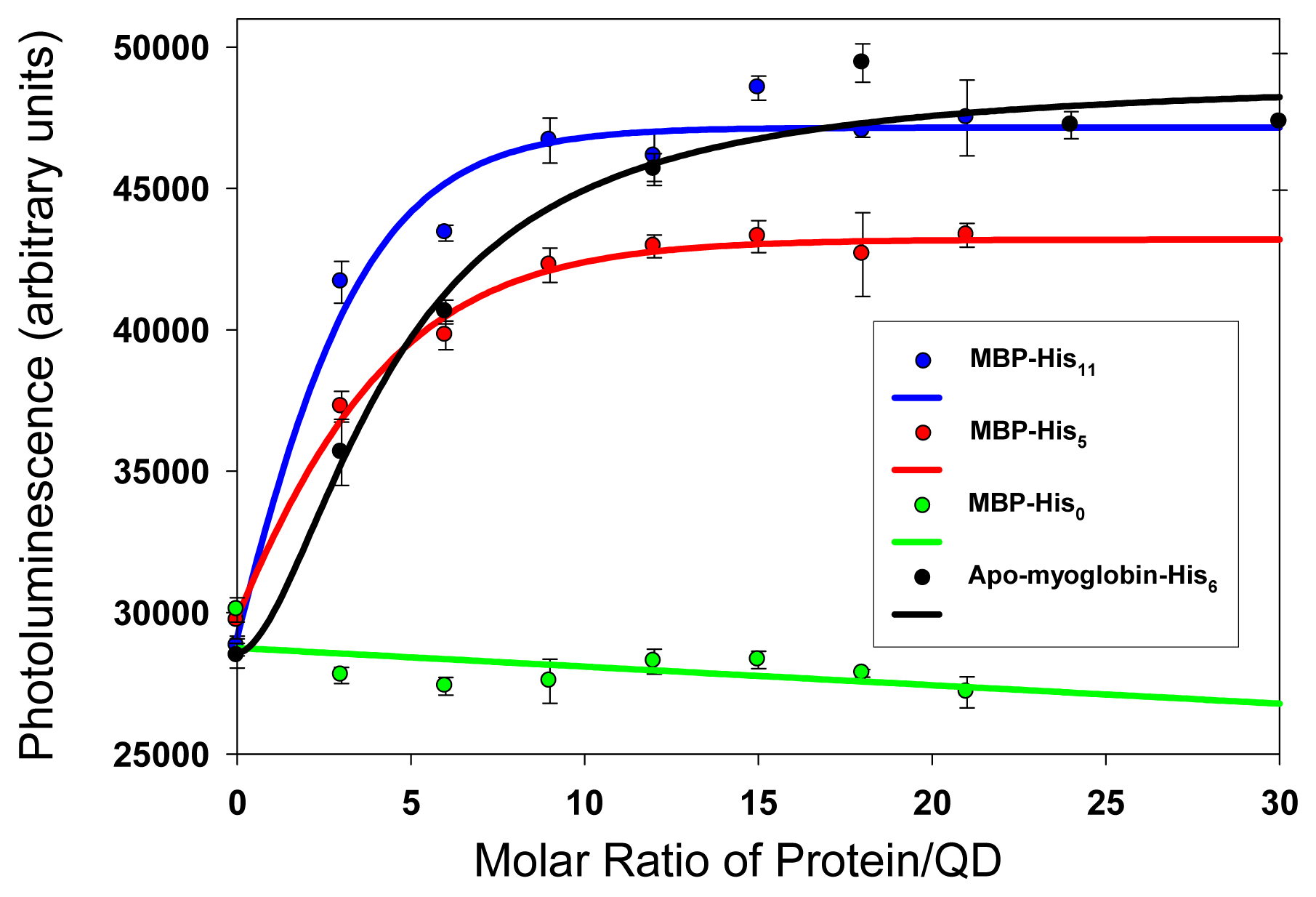
3. Bioconjugation of Quantum Dots
4. Generalized Probes
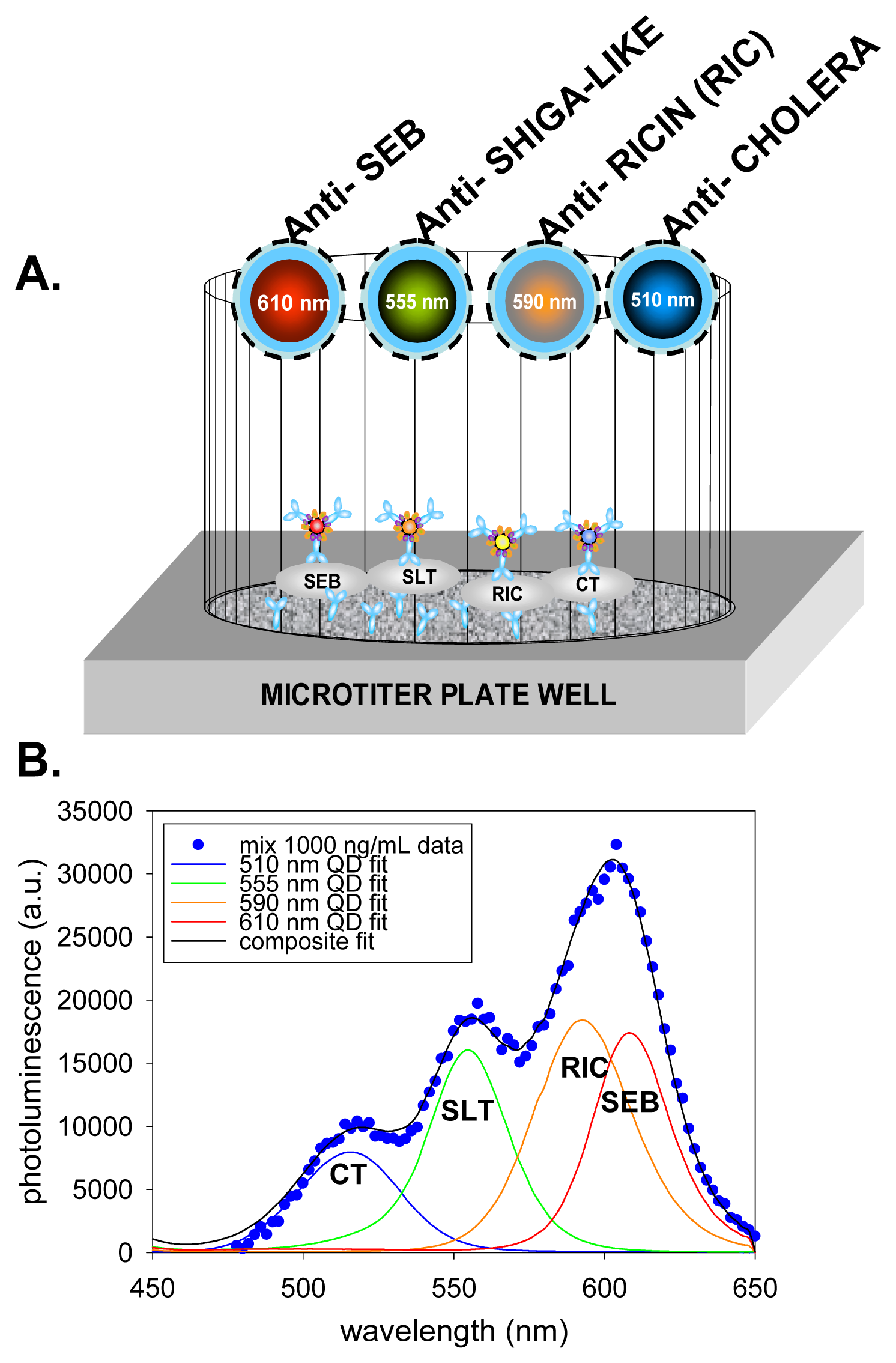
5. Immunoassays Using Quantum Dots
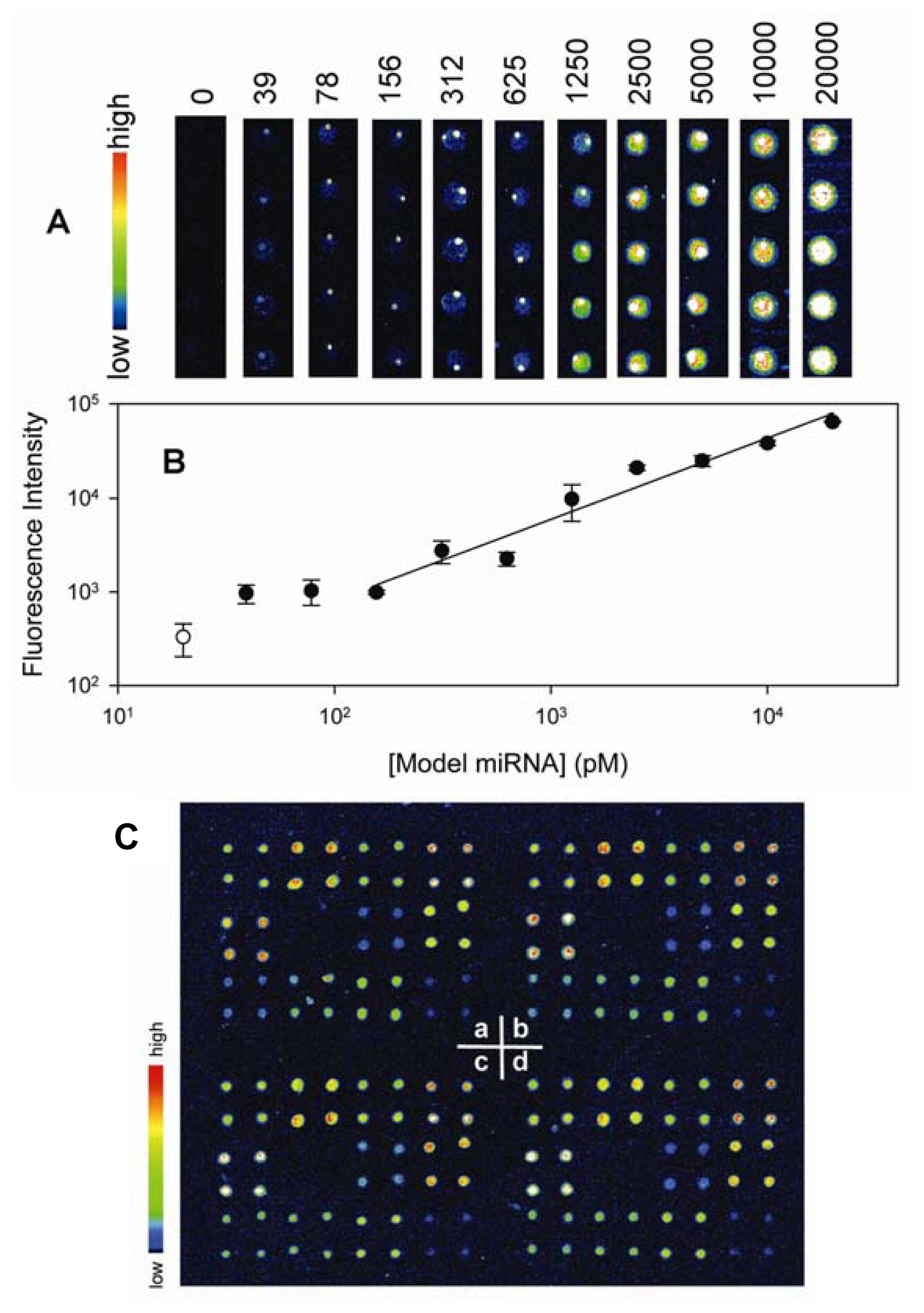
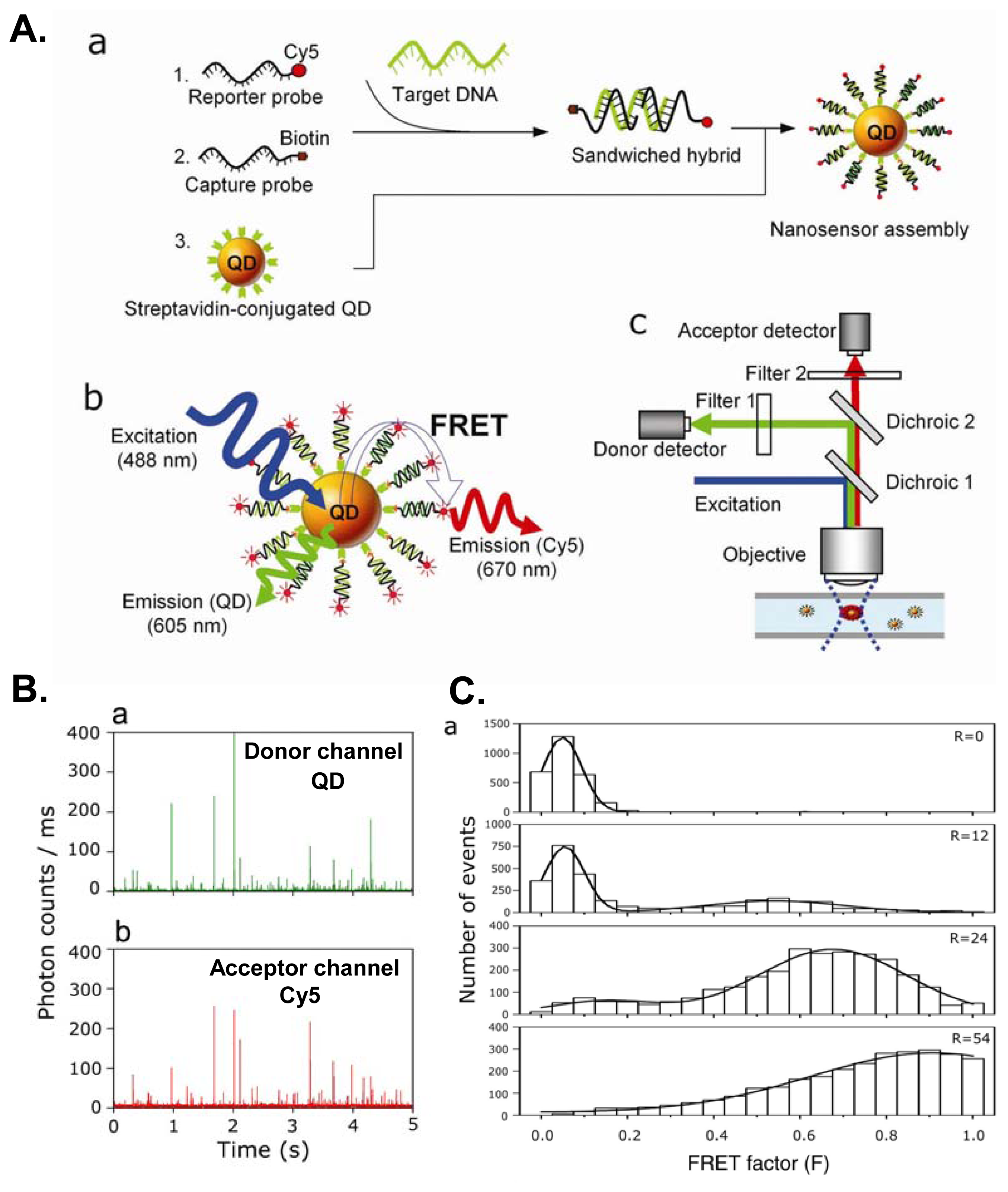
6. Nucleic Acid Detection
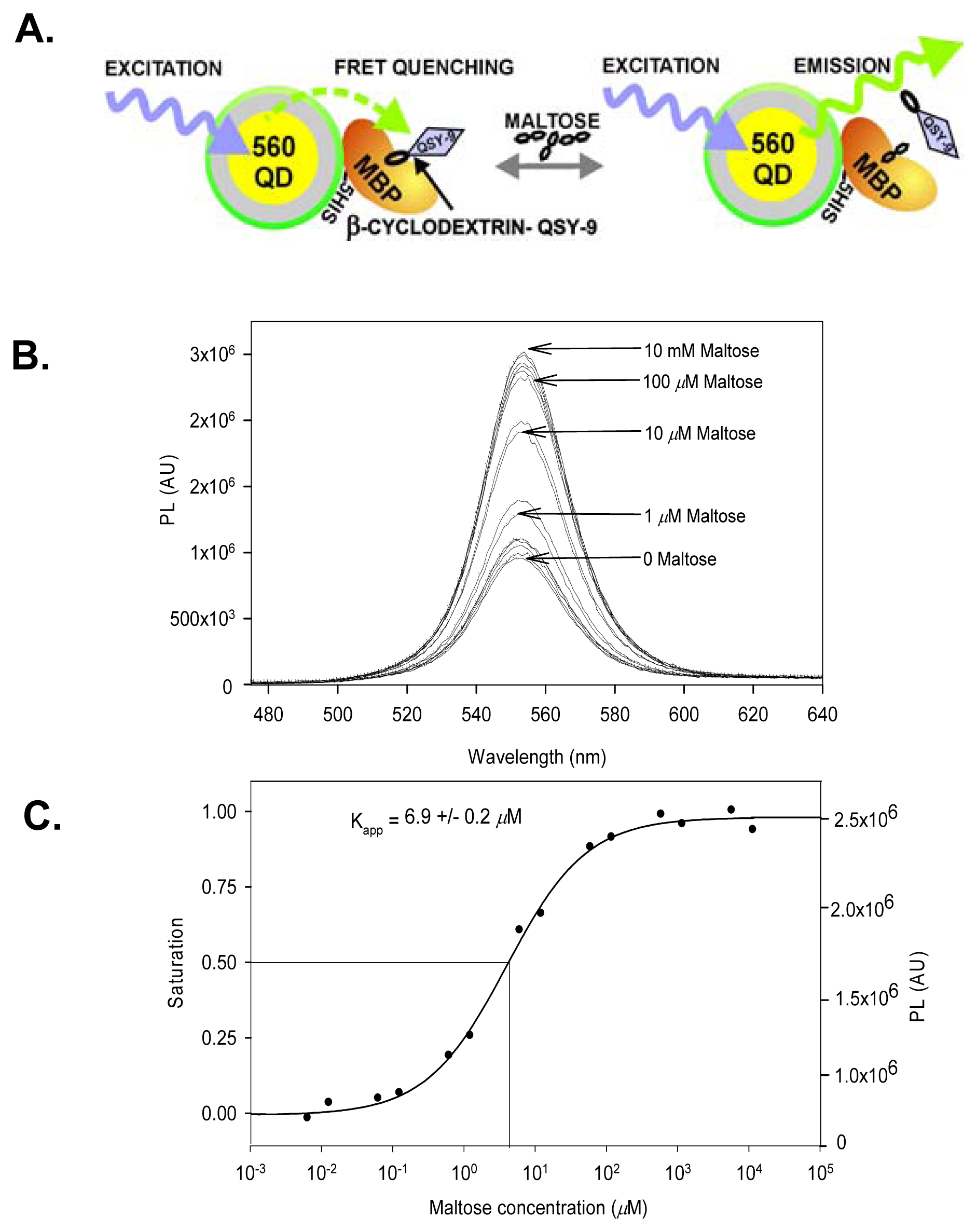
7. Sensing Based on FRET with Quantum Dot Bioconjugates
8. Other Quantum Dot Biosensing Modalities
9. Considerations When Using Quantum Dots for Biosensing
Acknowledgments
References and Notes
- Haugland, R.P. The Handbook A Guide to Fluorescent Probes and Labeling Technologies, Tenth Edition ed; Invitrogen Corporation; San Diego, 2005. [Google Scholar]
- Lakowicz, JR. Principles of Fluorescence Spectroscopy, Second Edition ed; Kluwer Academic/Plenum Publishers: New York, 1999. [Google Scholar]
- Giepmans, B.N.G.; Adams, S.R.; Ellisman, M.H.; Tsien, R.Y. The Fluorescent Toolbox for Assessing Protein Location and Function. Science 2006, 312, 217–224. [Google Scholar]
- Kricka, L.J. Stains, Labels and Detection Strategies for Nucleic Acids Assays. Annals of Clinical Biochemistry 2002, 39, 114–129. [Google Scholar]
- Chan, W.C.W.; Nie, S. Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection. Science 1998, 281, 2016–2018. [Google Scholar]
- Bruchez, M., Jr.; Moronne, M.; Gin, P.; Weiss, S.; Alivisatos, A.P. Semiconductor Nanocrystals as Fluorescent Biological Labels. Science 1998, 281, 2013–2016. [Google Scholar]
- Medintz, I.L.; Uyeda, H.T.; Goldman, E.R.; Mattoussi, H. Quantum Dot Bioconjugates for Imaging, Labelling and Sensing. Nature Materials 2005, 4, 435–446. [Google Scholar]
- Michalet, X.; Pinaud, F.F.; Bentolila, L.A.; Tsay, J.M.; Doose, S.; Li, J.J.; Sundaresan, G.; Wu, A.M.; Gambhir, S.S.; Weiss, S. Quantum Dots for Live Cells, In Vivo Imaging, and diagnostics. Science 2005, 307, 538–544. [Google Scholar]
- Alivisatos, A.P.; Gu, W.; Larabell, C.A. Quantum Dots as Cellular Probes. Ann. Rev. Biomed. Eng. 2005, 7, 55–76. [Google Scholar]
- Parak, W.J.; Pellegrino, T.; Plank, C. Labelling of Cells with Quantum Dots. Nanotech 2005, 16, R9–R25. [Google Scholar]
- Parak, W.J.; Boudreau, R.; Le Gros, M.; Gerion, D.; Zanchet, D.; Micheel, C.M.; Williams, S.C.; Alivisatos, A.P.; Larabell, C. Cell Motility and Metastatic Potential Studies Based on Quantum Dot Imaging of Phagokinetic Tracks. Adv. Mater. 2002, 14, 882–885. [Google Scholar]
- Parak, W.J.; Gerion, D.; Pellegrino, T.; Zanchet, D.; Micheel, C.; Williams, S.C.; Boudreau, R.; Le Gros, M.A.; Larabell, C.A.; Alivisatos, A.P. Biological Applications of Colloidal Nanocrystals. Nanotech. 2003, 14, R15–R27. [Google Scholar]
- Murray, C.B.; Norris, D.J.; Bawendi, M.G. Synthesis and Characterization of Nearly Monodisperse CdE (E = Sulfur, Selenium, Tellurium) Semiconductor Nanocrystallites. J. Am. Chem. Soc. 1993, 115, 8706–8715. [Google Scholar]
- Dabbousi, B.O.; Rodriguez-Viejo, J.; Mikulec, F.V.; Heine, J.R.; Mattoussi, H.; Ober, R.; Jensen, K.F.; Bawendi, M.G. (CdSe)ZnS Core-Shell Quantum Dots: Synthesis and Optical and Structural Characterization of a Size Series of Highly Luminescent Materials. J. Phys. Chem. B. 1997, 101, 9463–9475. [Google Scholar]
- Peng, X.; Schlamp, M.C.; Kadavanich, A.V.; Alivisatos, A.P. Epitaxial Growth of Highly Luminescent CdSe/CdS Core/Shell Nanocrystals with Photostability and Electronic Accessibility. J. Am. Chem. Soc. 1997, 119, 7019–7029. [Google Scholar]
- Murray, C.B.; Kagan, C.R.; Bawendi, M.G. Synthesis and Characterization of Monodisperse Nanocrystals and Close-Packed Nanocrystal Assemblies. Ann. Rev. Mater. Sci. 2000, 30, 545–610. [Google Scholar]
- Derfus, A.M.; Chan, W.C.W.; Bhatia, S.N. Probing the Cytotoxicity of Semiconductor Quantum Dots. NanoLett. 2004, 4, 11–18. [Google Scholar]
- Hines, M.A.; Guyot-Sionnest, P. Synthesis and Characterization of Strongly Luminescing ZnS-Capped CdSe Nanocrystals. J. Phys. Chem. 1996, 100, 468–471. [Google Scholar]
- Peng, Z.A.; Peng, X. Formation of High-Quality CdTe, CdSe, and CdS Nanocrystals Using CdO as Precursor. J. Am. Chem. Soc. 2001, 123, 183–184. [Google Scholar]
- Ozkan, M. Quantum Dots and Other Nanoparticles: What Can They Offer to Drug Discovery? Drug Discovery Today 2004, 9, 1065–1071. [Google Scholar]
- Murphy, C.J. Optical Sensing with Quantum Dots. Anal. Chem. 2002, 74, 520A–526A. [Google Scholar]
- Kippeny, T.; Swafford, L.A.; Rosenthal, S.J. Semiconductor Nanocrystals: A Powerful Visual Aid for Introducing the Particle in a Box. J. Chem. Ed. 2002, 79, 1094–1100. [Google Scholar]
- Alivisatos, A.P. The Use of Nanocrystals in Biological Detection. Nature Biotech. 2004, 22, 47–52. [Google Scholar]
- Striolo, A.; Ward, J.; Prausnitz, J.M.; Parak, W.J.; Zanchet, D.; Gerion, D.; Milliron, D.; Alivisatos, A.P. Molecular Weight, Osmotic Second Virial Coefficient, and Extinction Coefficient of Colloidal CdSe Nanocrystals. J. Phys. Chem. B. 2002, 106, 5500–5505. [Google Scholar]
- Leatherdale, C.A.; Woo, W.K.; Mikulec, F.V.; Bawendi, M.G. On the Absorption Cross Section of CdSe Nanocrystal Quantum Dots. J. Phys. Chem. B. 2002, 106, 7619–7622. [Google Scholar]
- Schrock, E.; duManoir, S.; Veldman, T.; Schoell, B.; Wienberg, J.; FergusonSmith, M.A.; Ning, Y.; Ledbetter, D.H.; BarAm, I.; Soenksen, D.; et al. Multicolor Spectral Karyotyping of Human Chromosomes. Science 1996, 273, 494–497. [Google Scholar]
- Bailey, R.E.; Smith, AM.; Nie, S.M. Quantum Dots in Biology and Medicine. Physica E-Low-Dimensional Systems & Nanostructures 2004, 25, 1–12. [Google Scholar]
- Pellegrino, T.; Kudera, S.; Liedl, T.; Javier, A.M.; Manna, L.; Parak, W.J. On the Development of Colloidal Nanoparticles Towards Multifunctional Structures and Their Possible Use for Biological Applications. Small 2005, 1, 48–63. [Google Scholar]
- Riegler, J.; Nann, T. Application of Luminescent Nanocrystals as Labels for Biological Molecules. Anal. Bioanal. Chem. 2004, 379, 913–919. [Google Scholar]
- Hermanson, G.T. Bioconjugate Techniques; Academic Press: San Diego, 1996. [Google Scholar]
- Zhang, C.Y.; Ma, H.; Nie, S.M.; Ding, Y.; Jin, L.; Chen, D.Y. Quantum Dot-Labeled Trichosanthin. Analyst 2000, 125, 1029–1031. [Google Scholar]
- Berti, L.; Xie, J.; Medintz, I.L.; Glazer, A.N.; Mathies, R.A. Energy Transfer Cassettes for Facile Labeling of Sequencing and PCR Primers. Anal. Biochem. 2001, 292, 188–197. [Google Scholar]
- Goldman, E.R.; Medintz, I.L.; Mattoussi, H. Luminescent Quantum Dots in Immunoassays. Anal. Bioanal. Chem. 2006, 384, 560–563. [Google Scholar]
- Mattoussi, H.; Mauro, J.M.; Goldman, E.R.; Anderson, G.P.; Sundar, V.C.; Mikulec, F.V.; Bawendi, M.G. Self-Assembly of CdSe-ZnS Quantum Dot Bioconjugates Using an Engineered Recombinant Protein. J. Am. Chem. Soc. 2000, 122, 12142–12150. [Google Scholar]
- Zhang, P. Investigation of Novel Quantum Dots/Proteins/Cellulose Bioconjugate using NSOM and Fluorescence. J. Fluorescence 2006, 16, 349–353. [Google Scholar]
- Slocik, J.M.; Moore, J.T.; Wright, D.W. Monoclonal Antibody Recognition of Histidine-Rich Peptide Encapsulated Nanoclusters. NanoLett. 2002, 2, 169–173. [Google Scholar]
- Sandros, M.G.; Gao, D.; Benson, D.E. A Modular Nanoparticle-Based System for Reagentless Small Molecule Biosensing. J. Am. Chem. Soc. 2005, 127, 12198–12199. [Google Scholar]
- Sandros, M.G.; Gao, D.; Gokdemir, C.; Benson, D.E. General, High-Affinity Approach for the Synthesis of Fluorophore Appended Protein Nanoparticle Assemblies. Chem. Comm. 2005, 22, 2832–2834. [Google Scholar]
- Sandros, M.G.; Shete, V.; Benson, D.E. Selective, Reversible, Reagentless Maltose Biosensing with Core-Shell Semiconducting Nanoparticles. The Analyst 2006, 131, 229–235. [Google Scholar]
- Medintz, I.L.; Clapp, A.R.; Mattoussi, H.; Goldman, E.R.; Fisher, B.; Mauro, J.M. Self-Assembled Nanoscale Biosensors Based on Quantum Dot FRET Donors. Nature Materials 2003, 2, 630–638. [Google Scholar]
- Ding, S.Y.; Rumbles, G.; Jones, M.; Tucker, M.P.; Nedeljkovic, J.; Simon, M.N.; Wall, J.S.; Himmel, M.E. Bioconjugation of (CdSe)ZnS Quantum Dots Using a Genetically Engineered Multiple Polyhistidine Tagged Cohesin/Dockerin Protein Polymer. Macromol. Mat. Eng. 2004, 289, 622–628. [Google Scholar]
- Ding, S.Y.; Jones, M.; Tucker, M.P.; Nedeljkovic, J.M.; Wall, J.; Simon, M.N.; Rumbles, G.; Himmel, M.E. Quantum Dot Molecules Assembled with Genetically Engineered Proteins. NanoLetters 2003, 3, 1581–1585. [Google Scholar]
- Pinaud, F.; King, D.; Moore, H.P.; Weiss, S. Bioactivation and Cell Targeting of Semiconductor CdSe/ZnS Nanocrystals with Phytochelatin-Related Peptides. J. Am. Chem. Soc. 2004, 126, 6115–6123. [Google Scholar]
- Tsay, J.M.; Doose, S.; Weiss, S. Rotational and Translational Diffusion of Peptide-Coated CdSe/CdS/ZnS Nanorods Studied by Fluorescence Correlation Spectroscopy. J. Am. Chem. Soc. 2006, 128, 1639–1647. [Google Scholar]
- Tsay, J.M.; Doose, S.; Pinaud, F.; Weiss, S. Enhancing the Photoluminescence of Peptide-Coated Nanocrystals with Shell Composition and UV Irradiation. J. Phys. Chem. B 2005, 109, 1669–1674. [Google Scholar]
- Ji, X.J.; Zheng, J.Y.; Xu, J.M.; Rastogi, V.K.; Cheng, T.C.; DeFrank, J.J.; Leblanc, R.M. (CdSe)ZnS Quantum Dots and Organophosphorus Hydrolase Bioconjugate as Biosensors for Detection of Paraoxon. J. Phys. Chem. B 2005, 109, 3793–3799. [Google Scholar]
- Constantine, C.A.; Gattas-Asfura, K.M.; Mello, S.V.; Crespo, G.; Rastogi, V.; Cheng, T.C.; De Frank, J.J.; Leblanc, R.M. Layer-by-Layer Films of Chitosan, Organophosphorus Hydrolase and Thioglycolic Acid-Capped CdSe Quantum Dots for the Detection of Paraoxon. J. Phys. Chem. B 2003, 107, 13762–13764. [Google Scholar]
- Constantine, C.A.; Gattas-Asfura, K.M.; Mello, S.V.; Crespo, G.; Rastogi, V.; Cheng, T.-C.; DeFrank, J.J.; Leblanc, R.M. Layer-by-Layer Biosensor Assembly Incorporating Functionalized Quantum Dots. Langmuir 2003, 19, 9863–9867. [Google Scholar]
- Goldman, E.R.; Mattoussi, H.M.; Anderson, G.P.; Medintz, I.L.; Mauro, J.M. Fluoroimmunoassays Using Antibody-Conjugated Quantum Dots. Methods Molecular Biology 2005, 303, 19–34. [Google Scholar]
- Goldman, E.R.; Anderson, G.P.; Tran, P.T.; Mattoussi, H.; Charles, P.T.; Mauro, J.M. Conjugation of Luminescent Quantum Dots with Antibodies Using an Engineered Adaptor Protein to Provide New Reagents for Fluoroimmunoassays. Anal. Chem. 2002, 74, 841–847. [Google Scholar]
- Goldman, E.R.; Balighian, E.D.; Kuno, M.K.; Labrenz, S.; Tran, P.T.; Anderson, G.P.; Mauro, J.M.; Mattoussi, H. Luminescent Quantum Dot-Adaptor Protein-Antibody Conjugates for Use in Fluoroimmunoassays. Phys. Stat. Sol. B. 2002, 229, 407–414. [Google Scholar]
- Goldman, E.R.; Balighian, E.D.; Mattoussi, H.; Kuno, M.K.; Mauro, J.M.; Tran, P.T.; Anderson, G.P. Avidin: A Natural Bridge for Quantum Dot-Antibody Conjugates. J. Am. Chem. Soc. 2002, 124, 6378–6382. [Google Scholar]
- Goldman, E.R.; Clapp, A.R.; Anderson, G.P.; Uyeda, H.T.; Mauro, J.M.; Medintz, I.L.; Mattoussi, H. Multiplexed Toxin Analysis Using Four Colors of Quantum Dot Fluororeagents. Anal. Chem. 2004, 76, 684–688. [Google Scholar]
- Goldman, E.R.; Medintz, I.L.; Hayhurst, A.; Anderson, G.P.; Mauro, J.M.; Iverson, B.L.; Georgiou, G.; Mattoussi, H. Self-Assembled Luminescent CdSe-ZnS Quantum Dot Bioconjugates Prepared Using Engineered Poly-Histidine Terminated Proteins. Analytica Chimica Acta 2005, 534, 63–67. [Google Scholar]
- Goldman, E.; Medintz, I.; Whitley, J.; Hayhurst, A.; Clapp, A.; Uyeda, H.; Deschamps, J.; Lassman, M.; Mattoussi, H. A Hybrid Quantum Dot-Antibody Fragment Fluorescence Resonance Energy Transfer-Based TNT Sensor. J. Am. Chem. Soc. 2005, 127, 6744–6751. [Google Scholar]
- Mamedova, N.N.; Kotov, N.A.; Rogach, A.L.; Studer, J. Albumin-CdTe Nanoparticle Bioconjugates: Preparation, Structure, and Interunit Energy Transfer with Antenna Effect. NanoLett. 2001, 1, 281–286. [Google Scholar]
- Hanaki, K.; Momo, A.; Oku, T.; Komoto, A.; Maenosono, S.; Yamaguchi, Y.; Yamamoto, K. Semiconductor Quantum Dot/Albumin Complex is a Long-Life and Highly Photostable Endosome Marker. Biochem. Biophys. Res. Comm. 2003, 302, 496–501. [Google Scholar]
- Wang, L.Y.; Wang, L.; Gao, F.; Yu, Z.Y.; Wu, Z.M. Application of Functionalized CdS Nanoparticles as Fluorescence Probe in the Determination of Nucleic Acids. Analyst 2002, 127, 977–980. [Google Scholar]
- Wang, L.Y.; Kan, X.W.; Zhang, M.C.; Zhu, C.Q.; Wang, L. Fluorescence for the Determination of Protein with Functionalized Nano-ZnS. Analyst 2002, 127, 1531–1534. [Google Scholar]
- Zhu, C.Q.; Zhao, D.H.; Chen, J.L.; Li, Y.X.; Wang, L.Y.; Wang, L.; Zhou, Y.Y.; Zhuo, S.J.; Wu, Y.Q. Application of L-Cysteine-Capped Nano-ZnS as a Fluorescence Probe for the Determination of Proteins. Anal. Bioanal. Chem. 2004, 378, 811–815. [Google Scholar]
- Lin, Z.; Cui, S.; Zhang, H.; Chen, Q.; Yang, B.; Su, X.; Zhang, J.; Jin, Q. Studies on Quantum Dots Synthesized in Aqueous Solution for Biological Labeling Via Electrostatic Interaction. Analytical Biochemistry 2003, 319, 239–243. [Google Scholar]
- Chen, X.D.; Wang, X.B.; Liu, L.; Yang, D.C.; Fan, L. Functionalized Semiconductor Nanocrystals for Ultrasensitive Detection of Peptides. Analytica Chimica Acta 2005, 542, 144–150. [Google Scholar]
- Liang, J.; Huang, S.; Zeng, D.; He, Z.; Ji, X.J.; Ai, X.; Yang, H. CdSe Quantum Dots as Luminescent Probes for Spironolactone Determination. Talanta 2006, 69, 126–130. [Google Scholar]
- Gattas-Asfura, K.A.; Leblanc, R.M. Peptide-Coated CdS Quantum Dots for the Optical Detection of Copper(II) and Silver(I). Chemical Comm. 2003, 21, 2684–2685. [Google Scholar]
- Bo, C.; Ping, Z. A New Determining Method of Copper(III) Ions at ng ml(-1) Levels Based on Quenching of the Water-Soluble Nanocrystals Fluorescence. Anal. Bioanal. Chem. 2005, 381, 986–992. [Google Scholar]
- Wang, L.; Xia, T.T.; Liu, J.S.; Wang, L.Y.; Chen, H.Q.; Dong, L.; Bian, G.R. Preparation and Application of a Novel Core/Shell Organic Nanoparticle as a Fluorescence Probe in the Selective Determination of Cr(VI). Spectrochimica Acta Part a-Molecular and Biomolecular Spectroscopy 2005, 62, 565–569. [Google Scholar]
- She, S.K.; Zhou, Y.Y.; Zhang, L.; Wang, L.Y.; Wang, L. Preparation of Fluorescent Polyvinyl Alcohol Keto-Derivatives Nanoparticles and Selective Determination of Chromium(VI). Spectrochimica Acta Part a-Molecular and Biomolecular Spectroscopy 2005, 62, 711–715. [Google Scholar]
- Jin, W.J.; Fernandez-Arguelles, M.T.; Costa-Fernandez, J.M.; Pereiro, R.; Sanz-Medel, A. Photoactivated Luminescent CdSe Quantum Dots as Sensitive Cyanide Probes in Aqueous Solutions. Chem. Comm. 2005, 7, 883–885. [Google Scholar]
- Wang, S.P.; Mamedova, N.; Kotov, N.A.; Chen, W.; Studer, J. Antigen/Antibody Immunocomplex from CdTe Nanoparticle Bioconjugates. NanoLetters 2002, 2, 817–822. [Google Scholar]
- Sun, B.Q.; Xie, W.Z.; Yi, G.S.; Chen, D.P.; Zhou, Y.X.; Cheng, J. Microminiaturized Immunoassays Using Quantum Dots as Fluorescent Label by Laser Confocal Scanning Fluorescence Detection. J. Immunological Methods 2001, 249, 85–89. [Google Scholar]
- Li, J.; Zhao, K.; Hong, X.; Yuan, H.; Ma, L.; Li, J.H.; Bai, Y.B.; Li, T.J. Prototype of Immunochromatographic Assay Strips Using Colloidal CdTe Nanocrystals as Biological Luminescent Label. Colloids and Surfaces B-Biointerfaces 2005, 40, 179–182. [Google Scholar]
- Zahavy, E.; Freeman, E.; Lustig, S.; Keysary, A.; Yitzhaki, S. Double Labeling and Simultaneous Detection of B- and T- Cells Using Fluorescent Nanocrystal (Q-Dots) in Paraffin-Embedded Tissues. J.Fluorescence 2005, 15, 661–665. [Google Scholar]
- Wang, H.Z.; Wang, H.Y.; Liang, R.Q.; Ruan, K.C. Detection of Tumor Marker CA125 in Ovarian Carcinoma Using Quantum Dots. Acta Biochimica Et Biophysica Sinica 2004, 36, 681–686. [Google Scholar]
- Colton, H.M.; Falls, J.G.; Ni, H.; Kwanyuen, P.; Creech, D.; McNeil, E.; Casey, W.M.; Hamilton, G.; Cariello, N.F. Visualization and Quantitation of Peroxisomes using Fluorescent Nanocrystals: Treatment of Rats and Monkeys with Fibrates and Detection in the Liver. Toxicological Sciences 2004, 80, 183–192. [Google Scholar]
- Stsiapura, V.; Sukhanova, A.; Artemyev, M.; Pluot, M.; Cohen, J.H.M.; Baranov, A.V.; Oleinikov, V.; Nabiev, I. Functionalized Nanocrystal-Tagged Fluorescent Polymer Beads: Synthesis, Physicochemical Characterization, and Immunolabeling Application. Analytical Biochemistry 2004, 334, 257–265. [Google Scholar]
- Jaiswal, J.K.; Goldman, E.R.; Mattoussi, H.; Simon, S.M. Use of Quantum Dots for Live Cell Imaging. Nature Methods 2004, 1, 73–78. [Google Scholar]
- Ornberg, R.L.; Harper, T.F.; Hongjian, L. Western Blot Analysis with Quantum Dot Fluorescence Technology: a Sensitive and Quantitative Method for Multiplexed Proteomics. Nature Methods 2005, 2, 79–81. [Google Scholar]
- Makrides, S.C.; Gasbarro, C.; Bello, J.M. Bioconjugation of Quantum Dot Luminescent Probes for Western Blot Analysis. Biotechniques 2005, 39, 501–506. [Google Scholar]
- Bakalova, R.; Zhelev, Z.; Ohba, H.; Baba, Y. Quantum Dot-Based Western Blot Technology for Ultrasensitive Detection of Tracer Proteins. J. Am. Chem. Soc. 2005, 127, 3928–9329. [Google Scholar]
- Zhelev, Z; Bakalova, R.; Ohba, H.; Jose, R.; Imai, Y.; Baba, Y. Uncoated, Broad Fluorescent, and Size-Homogeneous CdSe Quantum Dots for Bioanalyses. Anal. Chem. 2006, 78, 321–330. [Google Scholar]
- Geho, D.; Lahar, N.; Gurnani, P.; Huebschman, M.; Herrmann, P.; Espina, V.; Shi, A.; Wulfkuhle, J.; Garner, H.; Petricoin, E.; et al. Pegylated, Streptavidin-Conjugated Quantum dots are effective Detection Elements for Reverse-Phase Protein Microarrays. Bioconj. Chem. 2005, 16, 559–566. [Google Scholar]
- de Farias, P.M.A.; Santos, B.S.; de Menezes, F.D.; Ferreira, R.D.; Barias-Castro, M.L.; Castro, V.; Lima, P.R.M.; Fontes, A.; Cesar, C.L. Investigation of Red Blood Cell Antigens with Highly Fluorescent and Stable Semiconductor Quantum Dots. J. Biomedical Optics 2005, 10, 44023. [Google Scholar]
- Otsuka, Y.; Hanaki, K.; Zhao, J.Z.; Ohtsuki, R.; Toyooka, K.; Yoshikura, H.; Kuratsuji, T.; Yamamoto, K.; Kirikae, T. Detection of Mycobacterium bovis bacillus Calmette-Guerin Using Quantum Dot Immuno-Conjugates. Japanese J. Infect. Diseases 2004, 57, 183–184. [Google Scholar]
- Lee, L.Y.; Ong, S.L.; Hu, J.Y.; Ng, W.J.; Feng, Y.Y.; Tan, X.L.; Wong, S.W. Use of Semiconductor Quantum Dots for Photostable Immunofluorescence Labeling of Cryptosporidium parvum. App. Environ. Microbiol 2004, 70, 5732–5736. [Google Scholar]
- Zhu, L.; Ang, S.; Liu, W.T. Quantum Dots as a Novel Immunofluorescent Detection System for Cryptosporidium parvum and Giardia lamblia. App. Environ. Microbiol. 2004, 70, 597–598. [Google Scholar]
- Hahn, M.A.; Tabb, J.S.; Krauss, T.D. Detection of Single Bacterial Pathogens with Semiconductor Quantum Dots. Analyical Chemistry 2005, 77, 4861–4869. [Google Scholar]
- Yang, L.; Li, Y. Simultaneous Detection of Escherichia coli O157:H7 and Salmonella Typhimurium Using Quantum Dots as Fluorescence Labels. Analyst 2006, 131, 394–401. [Google Scholar]
- Zhang, Q.; Zhu, L.; Feng, H.H.; Ang, S.; Chau, F.S.; Liu, W.T. Microbial Detection in Microfluidic Devices Through Dual Staining of Quantum Dots-Labeled Immunoassay and RNA Hybridization. Analytica Chimica Acta 2006, 556, 171–177. [Google Scholar]
- Liu, W.T.; Zhu, L.; Qin, Q.W.; Zhang, Q.; Feng, H.H.; Ang, S. Microfluidic Device as a New Platform for Immunofluorescent Detection of Viruses. Lab on a Chip 2005, 5, 1327–1330. [Google Scholar]
- Bentzen, E.L.; House, F.; Utley, T.J.; Crowe, J.E.; Wright, D.W. Progression of Respiratory Syncytial Virus Infection Monitored by Fluorescent Quantum Dot Probes. NanoLett. 2005, 5, 591–595. [Google Scholar]
- Hoshino, A.; Fujioka, K.; Manabe, N.; Yamaya, S.; Goto, Y.; Yasuhara, M.; Yamamoto, K. Simultaneous Multicolor Detection System of the Single-Molecular Microbial Antigen with Total Internal Reflection Fluorescence Microscopy. Microbiology & Immunology 2005, 49, 461–470. [Google Scholar]
- Gerion, D.; Parak, W.J.; Williams, S.C.; Zanchet., D.; Micheel, C.M.; Alivisatos, A.P. Sorting Fluorescent Nanocrystals with DNA. J. Am. Chem. Soc. 2002, 124, 7070–7074. [Google Scholar]
- Gerion, D.; Chen, F.; Kannan, B.; Fu, A.; Parak, W.J.; Chen, D.J.; Majumdar, A.; Alivisatos, A.P. Room-Temperature Single-Nucleotide Polymorphism and Multiallele DNA Detection Using Fluorescent Nanocrystals and Microarrays. Anal. Chem. 2003, 75, 4766–4772. [Google Scholar]
- Shepard, J.R.E. Polychromatic Microarrays: Simultaneous Multicolor Array Hybridization of Eight Samples. Anal. Chem. 2006, 78, 2478–2486. [Google Scholar]
- Liang, R.Q.; Li, W.; Li, Y.; Tan, C.Y.; Li, J.X.; Jin, Y.X.; Ruan, K.C. An Oligonucleotide Microarray for microRNA Expression Analysis Based on Labeling RNA with Quantum Dot and NanogoldPprobe. Nucleic Acids Research 2005, 33, e17. [Google Scholar]
- Pathak, S.; Choi, S.K.; Arnheim, N.; Thompson, M.E. Hydroxylated Quantum Dots as Luminescent Probes for In Situ Hybridization. J. Am. Chem. Soc. 2001, 123, 4103–4104. [Google Scholar]
- Xu, H.X.; Sha, M.Y.; Wong, E.Y.; Uphoff, J.; Xu, Y.H.; Treadway, J.A.; Truong, A.; O'Brien, E.; Asquith, S.; Stubbins, M.; et al. Multiplexed SNP Genotyping Using the Qbead (TM) System: a Quantum Dot-Encoded Microsphere-Based Assay. Nucleic Acids Research 2003, 31, e43. [Google Scholar]
- Meissner, K.E.; Spillman, W.B., Jr. Quantum-Dot Tagged Microspheres for Fluid-Based Microarrays. Physica Status Solidi (c) 2003, 4, 1355–1359. [Google Scholar]
- Han, M.Y.; Gao, X.H.; Su, J.Z.; Nie, S. Quantum-Dot-Tagged Microbeads for Multiplexed Optical Coding of Biomolecules. Nature Biotech. 2001, 19, 631–635. [Google Scholar]
- Ho, Y.P.; Kung, M.C.; Yang, S.; Wang, T.H. Multiplexed Hybridization Detection with Multicolor Colocalization of Quantum Dot Nanoprobes. NanoLett. 2005, 5, 1693–1697. [Google Scholar]
- Chan, P.M.; Yuen, T.; Ruf, F.; Gonzalez-Maeso, J.; Sealfon, S.C. Method for Multiplex Cellular Detection of mRNAs Using Quantum Dot Fluorescent In Situ Hybridization. Nucleic Acids Research 2005, 33, e161. [Google Scholar]
- Xiao, Y.; Barker, P.E. Semiconductor Nanocrystal Probes for Human Metaphase Chromosomes. Nucleic Acids Research. 2004, 32, e161. [Google Scholar]
- Bakalova, R.; Zhelev, Z.; Ohba, H.; Baba, Y. Quantum Dot-Conjugated Hybridization Probes for Preliminary Screening of siRNA Sequences. J. Am. Chem. Soc. 2005, 127, 11328–11335. [Google Scholar]
- Crut, A.; Geron-Landre, B.; Bonnet, I.; Bonneau, S.; Desbiolles, P.; Escude, C. Detection of Single DNA Molecules by Multicolor Quantum-Dot End-Labeling. Nucleic Acids Research 2005, 33, e98. [Google Scholar]
- Sapsford, K.E.; Berti, L.; Medintz, I.L. Fluorescence Resonance Energy Transfer Concepts, Applications and Advances. Minerva Biotech 2005, 16, 253–279. [Google Scholar]
- Jares-Erijman, E.; Jovin, T. FRET Imaging. Nature Biotech. 2003, 21, 1387–1395. [Google Scholar]
- Miyawaki, A. Visualization of the Spatial and Temporal Dynamics of Intracellular Signaling. Dev. Cell 2003, 4, 295–305. [Google Scholar]
- Clapp, A.R.; Medintz, I.L.; Mattoussi, H. Forster Resonance Energy Transfer Investigations Using Quantum-Dot Fluorophores. ChemPhysChem 2006, 7, 47–57. [Google Scholar]
- Clapp, A.R.; Medintz, I.L.; Mauro, J.M.; Fisher, B.R.; Bawendi, M.G.; Mattoussi, H. Fluorescence Resonance Energy Transfer Between Quantum Dot Donors and Dye-Labeled Protein Acceptors. J. Am. Chem. Soc. 2004, 126, 301–310. [Google Scholar]
- Clapp, A.R.; Medintz, I.L.; Uyeda, H.T.; Fisher, B.R.; Goldman, E.R.; Bawendi, M.G.; Mattoussi, H. Quantum Dot-Based Multiplexed Fluorescence Resonance Energy Transfer. J. Am. Chem. Soc. 2005, 127, 18212–18221. [Google Scholar]
- Medintz, I.L.; Goldman, E.R.; Lassman, M.E.; Mauro, J.M. A Fluorescence Resonance Energy Transfer Sensor Based on Maltose Binding Protein. Bioconj. Chem. 2003, 14, 909–918. [Google Scholar]
- Levy, M.; Cater, S.F.; Ellington, A.D. Quantum-Dot Aptamer Beacons for the Detection of Proteins. Chembiochem 2005, 6, 2163–6. [Google Scholar]
- Medintz, I.L.; Clapp, A.R.; Melinger, J.S.; Deschamps, J.R.; Mattoussi, H. A Reagentless Biosensing Assembly Based on Quantum Dot-Donor Forster Resonance Energy Transfer. Adv. Mat. 2005, 17, 2450–2455. [Google Scholar]
- Patolsky, F.; Gill, R.; Weizmann, Y.; Mokari, T.; Banin, U; Willner, I. Lighting-Up the Dynamics of Telomerization and DNA Replication by CdSe-ZnS Quantum Dots. J. Am. Chem. Soc. 2003, 125, 13918–13919. [Google Scholar]
- Gill, R.; Willner, I.; Shweky, I.; Banin, U. Fluorescence Resonance Energy Transfer in CdSe/ZnS-DNA Conjugates: Probing hybridization and DNA cleavage. J. Phys. Chem. B 2005, 109, 23715–23719. [Google Scholar]
- Kim, J.H.; Morikis, D.; Ozkan, M. Adaptation of Inorganic Quantum Dots for Stable Molecular Beacons. Sensors Actuators B-Chemical 2004, 102, 315–319. [Google Scholar]
- Zhang, C.Y.; Yeh, H.C.; Kuroki, M.T.; Wang, T.H. Single-Quantum-Dot-Based DNA Nanosensor. Nature Materials 2005, 4, 826–831. [Google Scholar]
- Dubertret, B. Quantum Dots - DNA Detectives. Nature Materials 2005, 4, 797–798. [Google Scholar]
- Goldman, E.R.; Medintz, I.L.; Whitley, J.L.; Hayhurst, A.; Clapp, A.R.; Uyeda, H.T.; Deschamps, J.R.; Lassman, M.E.; Mattoussi, H. A Hybrid Quantum Dot-Antibody Fragment Fluorescence Resonance Energy Transfer-Based TNT Sensor. J. Am. Chem. Soc. 2005, 127, 6744–6751. [Google Scholar]
- Gueroui, Z.; Libchaber, A. Single-Molecule Measurements of Gold-Quenched Quantum Dots. Physical Review Letters 2004, 93, 166108. [Google Scholar]
- Chang, E.; Miller, J.S.; Sun, J.T.; Yu, W.W.; Colvin, V.L.; Drezek, R.; West, J.L. Protease-Activated Quantum Dot Probes. Biochem. Biophys. Res. Comm. 2005, 334, 1317–1321. [Google Scholar]
- Dyadyusha, L.; Yin, H.; Jaiswal, S.; Brown, T.; Baumberg, J.J.; Booy, F.P.; Melvin, T. Quenching of CdSe Quantum Dot Emission, a New Approach for Biosensing. Chem. Comm. 2005, 25, 3201–3203. [Google Scholar]
- Clapp, A.R.; Medintz, I.L.; Fisher, B.R.; Anderson, G.P.; Mattoussi, H. Can Luminescent Quantum Dots be Efficient Energy Acceptors with Organic Dye Donors? J. Am. Chem. Soc. 2005, 127, 1242–1250. [Google Scholar]
- So, M-K.; Xu, C.; Loening, A.M.; Gambir, S.S.; Rao., J. Self-Illuminating Quantum Dot Conjugates for In Vivo Imaging. Nature Biotech. 2006, 24, 339–343. [Google Scholar]
- Hansen, J.A.; Wang, J.; Kawde, A-N.; Xiang, Y.; Gothelf, K.V.; Collins, G. Quantum-Dot/Aptamer-Based Ultrasensitive Multi-Analyte Electrochemical Biosensor. J. Am. Chem. Soc. 2006, 128, 2228–2229. [Google Scholar]
- Wang, J.; Liu, G.; Merkoci, A. Electrochemical Coding Technology for Simultaneous Detection of Multiple DNA Targets. J. Am. Chem. Soc. 2003, 125, 3214–3215. [Google Scholar]
- De Lorimier, R.M.; Smith, J.J.; Dwyer, M.A.; Looger, L.L.; Sali, K.M.; Paavola, C.D.; Rizk, S.S.; Sadigov, S.; Conrad, D.W.; Loew, L.; et al. Construction of a Fluorescent Biosensor Family. Protein Science 2002, 11, 2655–2675. [Google Scholar]
- Ipe, B.I.; Lehnig, M.; Niemeyer, C.M. On the Generation of Free Radical Species From Quantum Dots. Small 2005, 1, 706–709. [Google Scholar]
- Clarke, S.J.; Hollmann, C.A.; Zhang, Z.H.; Suffern, D.; Bradforth, S.E.; Dimitrijevic, N.M.; Minarik, W.G.; Nadeau, J.L. Photophysics of Dopamine-Modified Quantum Dots and Effects on Biological Systems. Nat. Materials 2006, 5, 409–417. [Google Scholar]
- Niehren, S.; Kinzelbach, W.; Seeger, S.; Wolfrum, J. An All-Solid-State Flow Cytometer for Counting Fluorescent Microspheres. Anal. Chem. 1995, 67, 2666–2671. [Google Scholar]
- Morseman, J.P.; Moss, M.W.; Zoha, S.J.; Allnutt, F.C.T. PBXL-1: A New Fluorochrome Applied to Detection of Proteins on Membranes. Biotechniques 1999, 26, 559–563. [Google Scholar]
- Sapsford, K.E.; Soto, C.M.; Blum, A.S.; Chatterji, A.; Lin, T.; Johnson, J.E.; Ligler, F.S.; Ratna, B.R. A Cowpea Mosaic Virus Nanoscaffold for Multiplexed Antibody Conjugation: Application As An Immunoassay Tracer. Biosensors & Bioelectronics 2006, 21, 1668–1673. [Google Scholar]
- Medintz, IL.; Konnert, J.H.; Clapp, A.R.; Stanish, I.; Twigg, M.E.; Mattoussi, H.; Mauro, J.M.; Deschamps, J.R. A Fluorescence Resonance Energy Transfer Derived Structure of a Quantum Dot-Protein Bioconjugate Nanoassembly. P. N. A. S. U. S. A. 2004, 101, 9612–9617. [Google Scholar]
- Tsien, R.Y. The Green Fluorescent Protein. Ann. Rev. Bioch. 1998, 67, 509–544. [Google Scholar]
- Wu, X.; Liu, H.; Liu, J.; Haley, K.N.; Treadway, J.A.; Larson, J.P.; Ge, N.; Peale, F.; Bruchez, M.P. Immunofluorescent Labeling of Cancer Marker Her2 and Other Cellular Targets with Semiconductor Quantum Dots. Nature Biotech. 2003, 21, 41–46. [Google Scholar]
- Jaiswal, J.K.; Simon, S.M. Potentials and Pitfalls of Fluorescent Quantum Dots for Biological Imaging. Trends in Cell Biology 2004, 14, 497–504. [Google Scholar]
- Larson, D.R.; Zipfel, W.R.; Williams, R.M.; Clark, S.W.; Bruchez, M.P.; Wise, F.W.; Webb, W.W. Water-Soluble Quantum Dots for Multiphoton Fluorescence Imaging In Vivo. Science 2003, 300, 1434–1437. [Google Scholar]
- Lidke, D.S.; Arndt-Jovin, D.J. Imaging Takes a Quantum Leap. Physiology 2004, 19, 322–325. [Google Scholar]
- Kuno, M.; Fromm, D.P.; Hamann, H.F.; Gallagher, A.; Nesbitt, D.J. ‘On’/‘Off’ Fluorescence Intermittency of Single Semiconductor Quantum Dots. J. Chem. Phys. 2001, 115, 1028–1040. [Google Scholar]
- Wang, C.J.; Shim, M.; Guyot-Sionnest, P. Electrochromic Nanocrystal Quantum Dots. Science 2001, 291, 2390–2392. [Google Scholar]

| Property | Fluorophores | Quantum Dots | Ref. |
|---|---|---|---|
| Photophysical | |||
| Absorption spectra | Variable/narrow generally a mirror of the emission spectra | Broad spectra, steadily increases towards the UV from the first absorption band edge | [1,14,24,25] |
| Molar extinction coefficients | Variable, Generally < 200,000 M-1 cm-1 | High, 10-100X that of fluorophores | [1,14,24,25] |
| Emission spectra | Broad, asymmetric red-tailed emission | Narrow-full width at half-maximum 25-40 nm for CdSe core materials | [14] |
| Maturation time | Needed for fluorescent proteins | NA | [134] |
| Effective Stokes shifts | Generally < 100 nm | > 200 nm possible | [8,20] |
| Tunable emission | NA | Unique to QDs / can be size-tuned from the UV to IR | [8] |
| Quantum yield | Variable, low to high | Generally high, 0.2 to 0.7 in buffer depending upon surface coating | [1,8,20] |
| Fluorescent lifetime | Short < 5 ns | Long ∼ 10-20 ns or greater | [8,20] |
| Spectral range | Necessitates a different dye every 40-60 nm | UV-IR depending upon binary/ternary material Vis - CdSe | [1,8,14,20] |
| Photostability | Variable to poor | Excellent, strong resistance to photobleaching several orders of magnitude that of dyes | [135,136] |
| Multiphoton cross section | Variable to poor | Excellent >2-3 orders of magnitude that of dyes | [137] |
| Single-molecule capabilities | Variable | Excellent | [138] |
| FRET capabilities | Variable, mostly single donor- single acceptor configurations | Excellent donors, size tune emission to improve the overlap with an acceptor dye, single donor-multiple acceptor configurations possible | [109,110] |
| Multiplexing capabilities | Rare | Excellent, largely unexplored | [8,20] |
| Intermittency (blinking) | Negligible | Maybe problematic in isolated circumstances (single molecule tracking) | [8,20,139] |
| Chemical | |||
| Chemical resistance | Variable | Excellent | |
| Reactivity | Multiple reactivities commercially available | Limited conjugation chemistries available | [20] |
| Mono-valent attachment | Easy | Difficult | |
| Multi-valent attachment | Rare – mostly bis-functional | Good possibilities, can attach several molecules to QDs depending upon size | [20] |
| Other | |||
| Physical size | < 0.5 nm | 4 – 7 nm diameter for CdSe core material | [8] |
| Electrochromicity | Rare | Largely untapped | [140] |
| Cost effectiveness | Very good / multiple suppliers | Poor / 2 commercial suppliers | [1] |
© 2006 by MDPI ( http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Sapsford, K.E.; Pons, T.; Medintz, I.L.; Mattoussi, H. Biosensing with Luminescent Semiconductor Quantum Dots. Sensors 2006, 6, 925-953. https://doi.org/10.3390/s6080925
Sapsford KE, Pons T, Medintz IL, Mattoussi H. Biosensing with Luminescent Semiconductor Quantum Dots. Sensors. 2006; 6(8):925-953. https://doi.org/10.3390/s6080925
Chicago/Turabian StyleSapsford, Kim E., Thomas Pons, Igor L. Medintz, and Hedi Mattoussi. 2006. "Biosensing with Luminescent Semiconductor Quantum Dots" Sensors 6, no. 8: 925-953. https://doi.org/10.3390/s6080925
APA StyleSapsford, K. E., Pons, T., Medintz, I. L., & Mattoussi, H. (2006). Biosensing with Luminescent Semiconductor Quantum Dots. Sensors, 6(8), 925-953. https://doi.org/10.3390/s6080925




