Advanced Molecular Solutions for Cancer Therapy—The Good, the Bad, and the Ugly of the Biomarker Paradigm
- (1)
- it provides a simple molecular mechanistic explanation of cancerization in any human and animal regardless of race/strain, sex, age, or any other personal and environmental characteristic;
- (2)
- (3)
- it provides the reason for developing universal assays to detect the existing cancer of a particular form and/or estimating the chances of future cancerization for any person;
- (4)
- it stimulated the development of animal models and engineered cell cultures to mimic various forms of human cancer, validate their genetic etiology, and test gene therapy;
- (5)
- it is the basis of designing therapeutic solutions that target the molecular mechanisms of cancerization;
- (6)
- it supports the standardization of the oncological procedures by the National Comprehensive Cancer Network (e.g., [20]);
- (7)
- it has been adopted by the vast majority of genomic researchers (as of 6 January 2024, PubMed [21] listed 421,759 “cancer biomarker” and 957,483 “cancer genetic etiology” publications);
- (8)
- it benefits from the most generous research funding by public and private agencies and is of major interest for the pharma industry.
- (1)
- low diagnostic specificity owing to the large number of cancer forms harboring the same biomarker;
- (2)
- insufficient diagnostic sensitivity, numerous cases missing the alleged biomarker (e.g., [35]);
- (3)
- disconsiders major personal favoring factors of the patient, including race, sex, age, diet, and environmental factors, such as exposure to radiation, toxins and stress;
- (4)
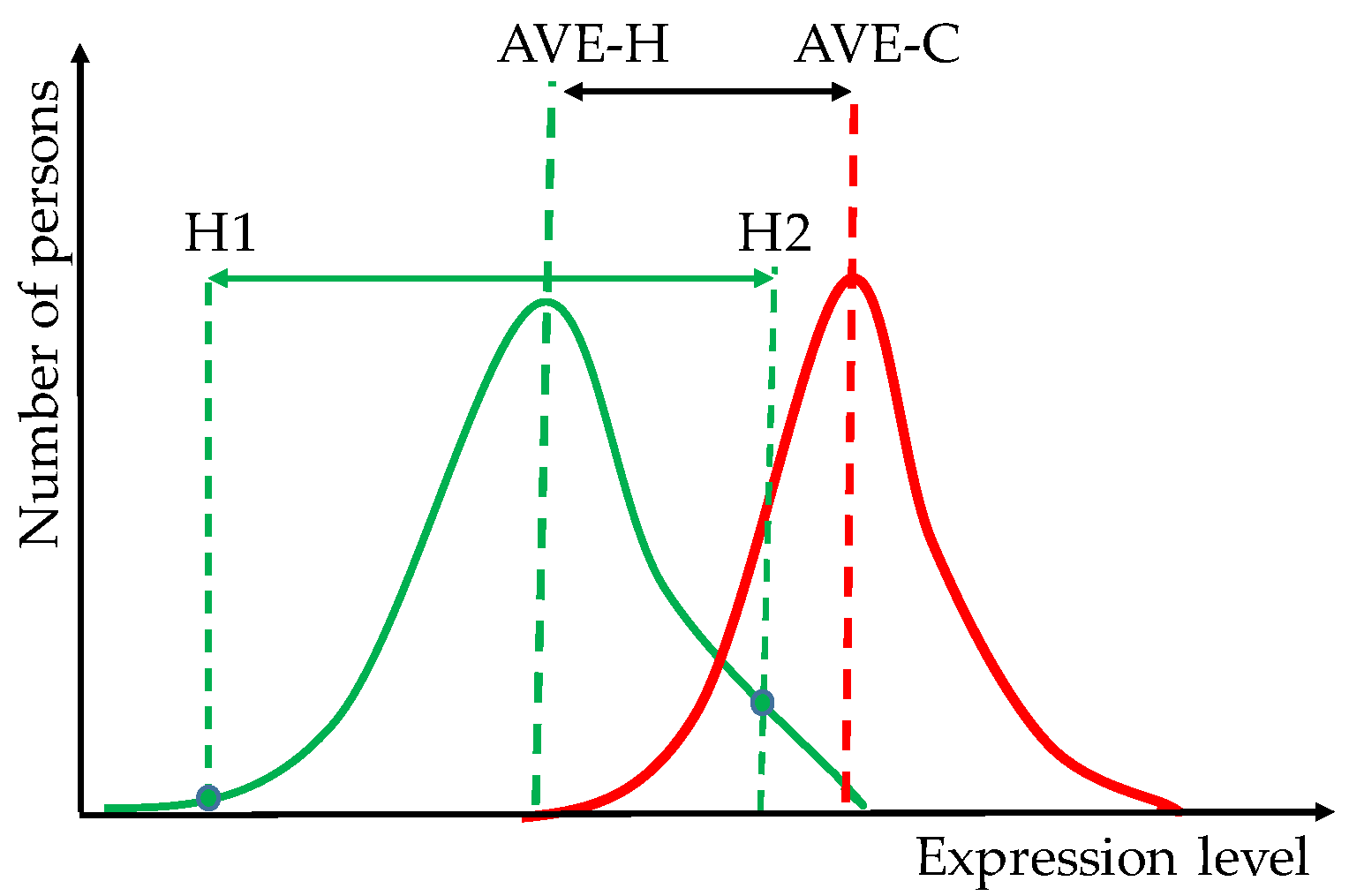
- differences between healthy persons might be larger than between the average healthy and the average cancer-stricken persons;
- (5)
- disregard of the contributions of the many other genes whose sequences and/or expression levels are altered (even in not repeatable combination) in the cancer of each individual;
- (6)
- BMP-based functional pathways do not discriminate with regard to race, sex, and age, do not change with the cancer progression and/or in response to external stimuli and treatments, and are reduced to unique gene networking;
- (7)
- it selects low cell players for gene therapy.
Conflicts of Interest
References
- Iacobas, D.A. Molecules at Play in Cancer. Curr. Issues Mol. Biol. 2023, 45, 2182–2185. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas (TCGA) Research Network. Available online: https://www.cancer.gov/tcga (accessed on 4 January 2024).
- Moço, P.D.; Farnós, O.; Sharon, D.; Kamen, A.A. Targeted Delivery of Chimeric Antigen Receptor into T Cells via CRISPR-Mediated Homology-Directed Repair with a Dual-AAV6 Transduction System. Curr. Issues Mol. Biol. 2023, 45, 7705–7720. [Google Scholar] [CrossRef]
- Wünsch, A.C.; Ries, E.; Heinzelmann, S.; Frabschka, A.; Wagner, P.C.; Rauch, T.; Koderer, C.; El-Mesery, M.; Volland, J.M.; Kübler, A.C.; et al. Metabolic Silencing via Methionine-Based Amino Acid Restriction in Head and Neck Cancer. Curr. Issues Mol. Biol. 2023, 45, 4557–4573. [Google Scholar] [CrossRef]
- Hwang, J.; Moon, H.; Kim, H.; Kim, K.-Y. Identification of a Novel ERK5 (MAPK7) Inhibitor, MHJ-627, and Verification of Its Potent Anticancer Efficacy in Cervical Cancer HeLa Cells. Curr. Issues Mol. Biol. 2023, 45, 6154–6169. [Google Scholar] [CrossRef]
- Albanna, H.; Gjoni, A.; Robinette, D.; Rodriguez, G.; Djambov, L.; Olson, M.E.; Hart, P.C. Activation of Adrenoceptor Alpha-2 (ADRA2A) Promotes Chemosensitization to Carboplatin in Ovarian Cancer Cell Lines. Curr. Issues Mol. Biol. 2023, 45, 9566–9578. [Google Scholar] [CrossRef]
- Savukaitytė, A.; Bartnykaitė, A.; Bekampytė, J.; Ugenskienė, R.; Juozaitytė, E. DDIT4 Downregulation by siRNA Approach Increases the Activity of Proteins Regulating Fatty Acid Metabolism upon Aspirin Treatment in Human Breast Cancer Cells. Curr. Issues Mol. Biol. 2023, 45, 4665–4674. [Google Scholar] [CrossRef]
- Filin, I.Y.; Mayasin, Y.P.; Kharisova, C.B.; Gorodilova, A.V.; Chulpanova, D.S.; Kitaeva, K.V.; Rizvanov, A.A.; Solovyeva, V.V. T-Lymphocytes Activated by Dendritic Cells Loaded by Tumor-Derived Vesicles Decrease Viability of Melanoma Cells In Vitro. Curr. Issues Mol. Biol. 2023, 45, 7827–7841. [Google Scholar] [CrossRef] [PubMed]
- Chatrath, A.; Przanowska, R.; Kiran, S.; Su, Z.; Saha, S.; Wilson, B.; Tsunematsu, T.; Ahn, J.H.; Lee, K.Y.; Paulsen, T.; et al. The pan-cancer landscape of prognostic germline variants in 10,582 patients. Genome Med. 2020, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Invitae Multi-Cancer Panel. Available online: https://www.invitae.com/us/providers/test-catalog/test-01101 (accessed on 11 December 2023).
- nCounter® PanCancer IO 360TM Panel. Available online: https://nanostring.com/products/ncounter-assays-panels/oncology/pancancer-io-360/ (accessed on 11 December 2023).
- TissueScan™ Cancer and Normal Tissue cDNA Arrays. Available online: https://www.origene.com/products/tissues/tissuescan (accessed on 12 November 2023).
- Vellekoop, H.; Huygens, S.; Versteegh, M.; Szilberhorn, L.; Zelei, T.; Nagy, B.; Koleva-Kolarova, R.; Tsiachristas, A.; Wordsworth, S.; Rutten-van Mölken, M. Guidance for the harmonisation and improvement of economic evaluations of personalised medicine. Pharmacoeconomics 2021, 39, 771–788. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wong, N.C.B.; Wang, Y.; Zemlyanska, Y.; Butani, D.; Virabhak, S.; Matchar, D.B.; Prapinvanich, T.; Teerawattananon, Y. Mapping the value for money of precision medicine: A systematic literature review and meta-analysis. Front. Public Health 2023, 11, 1151504. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Casarini, I.; Cobo, M.; Faivre-Finn, C.; Hegi-Johnson, F.; Lu, S.; Özgüroğlu, M.; Ramalingam, S.S. Targeted treatment for unresectable EGFR mutation-positive stage III non-small cell lung cancer: Emerging evidence and future perspectives. Lung Cancer 2023, 187, 107414. [Google Scholar] [CrossRef] [PubMed]
- Lutgendorf, S.K.; Telles, R.M.; Whitney, B.; Thaker, P.H.; Slavich, G.M.; Goodheart, M.J.; Penedo, F.J.; Noble, A.E.; Cole, S.W.; Sood, A.K.; et al. The biology of hope: Inflammatory and neuroendocrine profiles in ovarian cancer patients. Brain Behav. Immun. 2023, 116, 362–369. [Google Scholar] [CrossRef]
- QIAGEN Ingenuity Pathway Analysis (QIAGEN IPA). Available online: https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ (accessed on 7 January 2024).
- Database for Annotation, Visualization and Integrated Discovery (DAVID). Available online: https://david.ncifcrf.gov (accessed on 7 January 2024).
- Kyoto Encyclopedia of Genes and Genomes. Wiring Diagrams of Molecular Interactions, Reactions and Relations. Available online: https://www.genome.jp/kegg/pathway.html (accessed on 7 January 2024).
- Schaeffer, E.M.; Srinivas, S.; Adra, N.; An, Y.; Barocas, D.; Bitting, R.; Bryce, A.; Chapin, B.; Cheng, H.H.; D’Amico, A.V.; et al. Prostate Cancer, Version 4.2023, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2023, 21, 1067–1096. [Google Scholar] [CrossRef]
- NIH National Library of Medicine/PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/?term=cancer+biomarker&sort=date (accessed on 3 January 2024).
- NIH-National Cancer Institute Genomic Data Commons Data Portal. Available online: https://portal.gdc.cancer.gov (accessed on 29 November 2023).
- Ren, A.H.; Fiala, C.A.; Diamandis, E.P.; Kulasingam, V. Pitfalls in Cancer Biomarker Discovery and Validation with Emphasis on Circulating Tumor DNA. Cancer Epidemiol Biomark. Prev. 2020, 29, 2568–2574. [Google Scholar] [CrossRef]
- Kulac, I.; Roudier, M.P.; Haffner, M.C. Molecular Pathology of Prostate Cancer. Surg. Pathol. Clin. 2021, 14, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Tolkach, Y.; Kristiansen, G. The Heterogeneity of Prostate Cancer: A Practical Approach. Pathobiology 2018, 85, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.-M.; Zhang, M.; Wood, C.G.; Pisters, L.L. Stem Cell Theory of Cancer: Origin of Tumor Heterogeneity and Plasticity. Cancers 2021, 13, 4006. [Google Scholar] [CrossRef]
- Berglund, E.; Maaskola, J.; Schultz, N.; Friedrich, S.; Marklund, M.; Bergenstråhle, J.; Tarish, F.; Tanoglidi, A.; Vickovic, S.; Larsson, L.; et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018, 9, 2419. [Google Scholar] [CrossRef]
- Brady, L.; Kriner, M.; Coleman, I.; Morrissey, C.; Roudier, M.; True, L.D.; Gulati, R.; Plymate, S.R.; Zhou, Z.; Birditt, B.; et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat. Commun. 2021, 12, 1426. [Google Scholar] [CrossRef]
- Iacobas, S.; Iacobas, D.A. A Personalized Genomics Approach of the Prostate Cancer. Cells 2021, 10, 1644. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Obiomon, E.A.; Iacobas, S. Genomic Fabrics of the Excretory System’s Functional Pathways Remodeled in Clear Cell Renal Cell Carcinoma. Curr. Issues Mol. Biol. 2023, 45, 9471–9499. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Weber, Z.; San Lucas, F.A.; Deshpande, A.; Jakubek, Y.A.; Sulaiman, R.; Fagerness, M.; Flier, N.; Sulaiman, J.; Davis, C.M.; et al. Assessing inter-component heterogeneity of biphasic uterine carcinosarcomas. Gynecol Oncol. 2018, 151, 243–249. [Google Scholar] [CrossRef]
- Fujimoto, H.; Saito, Y.; Ohuchida, K.; Kawakami, E.; Fujiki, S.; Watanabe, T.; Ono, R.; Kaneko, A.; Takagi, S.; Najima, Y.; et al. Deregulated Mucosal Immune Surveillance through Gut-Associated Regulatory T Cells and PD-1+ T Cells in Human Colorectal Cancer. J. Immunol. 2018, 200, 3291–3303. [Google Scholar] [CrossRef]
- Yang, C.; Gong, J.; Xu, W.; Liu, Z.; Cui, D. Next-generation sequencing identified somatic alterations that may underlie the etiology of Chinese papillary thyroid carcinoma. Eur. J. Cancer Prev. 2023, 32, 264–274. [Google Scholar] [CrossRef]
- Clark, D.J.; Dhanasekaran, S.M.; Petralia, F.; Pan, J.; Song, X.; Hu, Y.; da Veiga Leprevost, F.; Reva, B.; Lih, T.M.; Chang, H.Y.; et al. Clinical Proteomic Tumor Analysis Consortium. Integrated Proteogenomic Characterization of Clear Cell Renal Cell Carcinoma. Cell 2019, 179, 964–983.e31. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, L.; Fang, C.; Li, C.; Zhang, L. Factors influencing the diagnostic and prognostic values of circulating tumor cells in breast cancer: A meta-analysis of 8,935 patients. Front Oncol. 2023, 13, 1272788. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacobas, D.A. Advanced Molecular Solutions for Cancer Therapy—The Good, the Bad, and the Ugly of the Biomarker Paradigm. Curr. Issues Mol. Biol. 2024, 46, 1694-1699. https://doi.org/10.3390/cimb46030109
Iacobas DA. Advanced Molecular Solutions for Cancer Therapy—The Good, the Bad, and the Ugly of the Biomarker Paradigm. Current Issues in Molecular Biology. 2024; 46(3):1694-1699. https://doi.org/10.3390/cimb46030109
Chicago/Turabian StyleIacobas, Dumitru Andrei. 2024. "Advanced Molecular Solutions for Cancer Therapy—The Good, the Bad, and the Ugly of the Biomarker Paradigm" Current Issues in Molecular Biology 46, no. 3: 1694-1699. https://doi.org/10.3390/cimb46030109




