In Vitro Assessment of Marine Bacillus for Use as Livestock Probiotics
Abstract
:1. Introduction
2. Results
2.1. Molecular Fingerprinting and Growth of Marine Bacillus
2.2. Sporulation Efficiency
2.3. Growth and Spore Germination under Anaerobic Conditions
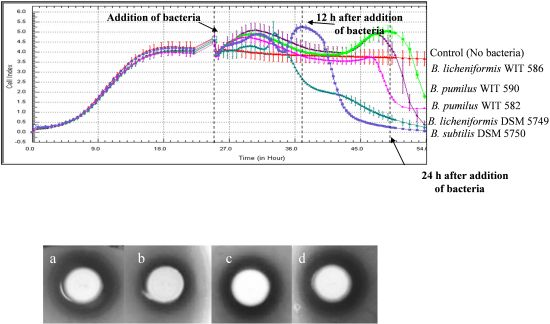
2.4. Antimicrobial Activity on Solid Media

| Test strain | E. coli | Salmonella | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| K88 | F18ab | F2S2 | F3P3 | F1L3 | F15OF3 | Typhimurium DT104 (DPC 6046) | Typhimurium PT12 (DPC 6465) | Typhimurium DT17 (WIT 396) | Typhimurium DT104 (WIT 387) | Derby (WIT 411) | |
| B. pumilus WIT 582 | + | +++ | ++ | ++ | ++ | + | ++ | + | + | ++ | ++ |
| B. pumilus WIT 584 | - | - | - | ++ | - | +/− | - | - | - | - | - |
| B. licheniformis WIT 586 | + | + | + | ++ | ++ | ++ | +/− | +/− | - | - | +/− |
| B. pumilus WIT 588 | +++ | +++ | ++ | +++ | +++ | ++ | ++ | +/− | - | - | - |
| B. pumilus WIT 590 | - | - | - | - | - | - | +/− | - | - | - | - |
| B. pumilus WIT 592 | - | - | - | - | - | - | - | +/− | - | - | - |
| B. licheniformis DSM 5749 2 | - | - | - | - | - | - | +/− | ++ | - | - | - |
| B. subtilis DSM 5750 2 | - | - | - | - | - | - | - | - | - | - | - |

2.5. Antimicrobial Activity in Liquid Culture
2.6. Bile Tolerance and Resistance to Simulated Gastrointestinal Conditions

| Test strain | Swarming motility 1 | Bile tolerance 2 | Adherence to intestinal epithelial cells 3 |
|---|---|---|---|
| B. pumilus WIT 582 | + | 1% | 17.4 ± 8.3 a |
| B. pumilus WIT 584 | − | <0.3% | 12.1 ± 10.6 a |
| B. licheniformis WIT 586 | + | 1% | 11.9 ± 5.1 a |
| B. pumilus WIT 588 | − | 1% 4 | 9.3 ± 4.0 a |
| B. pumilus WIT 590 | − | <0.3% | 11.0 ± 10.5 a |
| B. pumilus WIT 592 | + | <0.3% | 268.4 ± 239.7 b |
| B. licheniformis DSM 5749 | + | 2% 4 | 116.5 ± 79.9 b |
| B. subtilis DSM 5750 | + | 1% 4 | 196.6 ± 87.1 b |
| Lb. rhamnosus GG | ND 5 | ND | 705.6 ± 182.8 b |
2.7. Biofilm Formation, Swarming Motility and Adherence to Intestinal Epithelial Cells


2.8. In Vitro Safety Testing of Bacillus Isolates
2.8.1. Hemolytic Activity, Antibiotic Resistance and Presence of Enterotoxin Genes
| Test strain | Viability of intestinal epithelial cells (%) 1 | Hemolysis 2 |
|---|---|---|
| B. pumilus WIT 582 | 65.9 ± 3 | β |
| B. pumilus WIT 584 | 53 ± 10 | very weak β |
| B. licheniformis WIT 586 | 80.7 ± 4 | β |
| B. pumilus WIT 588 | 69.8 ± 3 | γ |
| B. pumilus WIT 590 | 86.2 ± 7 | very weak β |
| B. pumilus WIT 592 | 91.2 ± 6 | weak β |
| B. licheniformis DSM 5749 | 74.9 ± 6 | γ |
| B. subtilis DSM 5750 | 92.4 ± 10 | γ |
| Lb. rhamnosus GG | 68.6 ± 5 | ND |
2.8.2. Cytotoxic Activity against Intestinal Epithelial Cells

3. Discussion
4. Experimental Section
4.1. Bacterial Strains and Culture Conditions and Chemicals
4.2. Determination of Time Taken to Sporulate and Sporulation Efficiency and Preparation of Spores
4.3. Evaluation of Bacillus Growth and Spore Germination under Anaerobic Conditions
4.4. Assessment of Antimicrobial Activity of Bacillus Isolates on Solid Media
4.5. Assessment of Antimicrobial Activity of Bacillus Isolates in Liquid Culture
4.6. Bile, Salt Tolerance and Gastric and Ileum Juice Assays
4.7. Determination of Biofilm Formation and Swarming Motility
4.8. Evaluation of Adherence to Intestinal Epithelial Cells
4.8.1. Cell Culture
4.8.2. Assessment of Bacterial Growth in Cell Culture Medium
4.8.3. Adherence Assays

4.9. Analysis of Virulence Factors and Antibiotic Resistance
4.10. Evaluation of Cytotoxicity against Intestinal Epithelial Cells

4.11. Statistical Analysis
5. Conclusions
Supplementary Files
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pineiro, M.; Stanton, C. Probiotic bacteria: Legislative framework requirements to evidence basis. J. Nutr. 2007, 137, 850S–853S. [Google Scholar]
- Gaggìa, F.; Mattarelli, P.; Biavati, B. Probiotics and prebiotics in animal feeding for safe food production. Int. J. Food Microbiol. 2010, 141, S15–S28. [Google Scholar] [CrossRef]
- Balcázar, J.L.; Blas, I.D.; Ruiz-Zarzuela, I.; Cunningham, D.; Vendrell, D.; Múzquiz, J.L. The role of probiotics in aquaculture. Vet. Microbiol. 2006, 114, 173–186. [Google Scholar]
- Commission of the European Communities. REGULATION (EC) No 1831/2003 OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL of 22 September 2003 on additives for use in animal nutrition. Off. J. Eur. Union 2003, 268, 29–43. [Google Scholar]
- European Food Safety Authority (EFSA). Guidance for the preparation of dossiers for zootechnical additives. EFSA J. 2012, 10, 2536. [Google Scholar]
- Quigley, E.M. Prebiotics and probiotics; modifying and mining the microbiota. Pharmacol. Res. 2010, 61, 213–218. [Google Scholar] [CrossRef]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73, 386S–392S. [Google Scholar]
- Casula, G.; Cutting, S.M. Bacillus probiotics: Spore germination in the gastrointestinal tract. Appl. Environ. Microbiol. 2002, 68, 2344–2352. [Google Scholar] [CrossRef]
- Tam, N.K.M.; Uyen, N.Q.; Hong, H.A.; Duc, L.H.; Hoa, T.T.; Serra, C.R.; Henriques, A.O.; Cutting, S.M. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 2006, 188, 2692–2700. [Google Scholar] [CrossRef]
- EFSA. Scientific opinion on the modification to the formulation of GalliPro® and compatibility with formic acid. EFSA J. 2011, 9, 2112. [Google Scholar]
- Report of the Scientific Committee on Animal Nutrition on product Toyocerin® for use as feed additive. Available online: http://ec.europa.eu/food/fs/sc/scan/out72_en.pdf (accessed on 20 November 2013).
- Report of the Scientific Committee on Animal Nutrition on product Bioplus 2B® for use as feed additive. Available online: http://ec.europa.eu/food/fs/sc/scan/out49_en.pdf (accessed on 20 November 2013).
- EFSA. Scientific Opinion on the safety and efficacy of Calsporin® (Bacillus subtilis) for turkeys for fattening, ducks, geese, pigeons and other game birds for meat production, ducks, geese, pigeons, game birds, ornamental and sporting birds for rearing to point of lay, turkeys reared for breeding and chickens reared for laying. EFSA J. 2010, 8, 1867. [Google Scholar]
- Commission of the European Communities. COMMISSION REGULATION (EC) No 1292/2008 of 18 December 2008 concerning the authorisation of Bacillus amyloliquefaciens CECT 5940 (Ecobiol and Ecobiol plus) as a feed additive. Off. J. Eur. Union 2008, 340, 36–37. [Google Scholar]
- EFSA. Scientific opinion on the safety and efficacy of Bacillus subtilis PB6 (Bacillus subtilis) as a feed additive for chickens for fattening. EFSA J. 2009, 7, 1314. [Google Scholar]
- EFSA. Scientific Opinion on Animavit® (Bacillus subtilis CBS 117162) as feed additive for piglets and pigs for fattening. EFSA J. 2011, 9, 2375. [Google Scholar]
- Dobson, A.; Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocin production as a probiotic trait? Appl. Environ. Microbiol. 2012, 78, 1–6. [Google Scholar] [CrossRef]
- Penesyan, A.; Kjelleberg, S.; Egan, S. Development of novel drugs from marine surface-associated microorganisms. Mar. Drugs 2010, 8, 438–459. [Google Scholar] [CrossRef]
- Cutting, S.M. Bacillus probiotics. Food Microbiol. 2011, 28, 214–220. [Google Scholar] [CrossRef]
- Prieto, M.L.; O’Sullivan, L.; Tan, S.P.; McLoughlin, P.; Hughes, H.; O’Connor, P.M.; Cotter, P.D.; Lawlor, P.G.; Gardiner, G.E. Assessment of the bacteriocinogenic potential of marine bacteria reveals lichenicidin production by seaweed-derived Bacillus spp. Mar. Drugs 2012, 10, 2280–2299. [Google Scholar] [CrossRef]
- EFSA. Report of the task force on zoonoses data collection on the analysis of the baseline survey on the prevalence of Salmonella in slaughter pigs, Part A. EFSA J. 2008, 135, 1–111. [Google Scholar]
- Barbosa, T.M.; Serra, C.R.; Henriques, A.O. Gut Sporeformers. In Bacterial Spore Formers; Ricca, E., Henriques, A.O., Cutting, S.M., Eds.; Horizon Biosciences: Norfolk, UK, 2004. [Google Scholar]
- Fraser, G.M.; Hughes, C. Swarming motility. Curr. Opin. Microbiol. 1999, 2, 630–635. [Google Scholar] [CrossRef]
- EFSA. Technical guidance prepared by the Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) on the update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. EFSA J. 2008, 732, 1–15. [Google Scholar]
- Fakhry, S.; Sorrentini, I.; Ricca, E.; de Felice, M.; Baccigalupi, L. Characterization of spore forming bacilli isolated from the human gastrointestinal tract. J. Appl. Microbiol. 2008, 105, 2178–2186. [Google Scholar] [CrossRef]
- Ke, N.; Wang, X.; Xu, X.; Abassi, Y.A. The xCELLigence System for Real-time and Label-free Monitoring of Cell Viability. In Mammalian Cell Viability; Springer: San Diego, CA, USA, 2011; pp. 33–43. [Google Scholar]
- Guo, X.; Li, D.; Lu, W.; Piao, X.; Chen, X. Screening of Bacillus strains as potential probiotics and subsequent confirmation of the in vivo effectiveness of Bacillus subtilis MA139 in pigs. Antonie Van Leeuwenhoek 2006, 90, 139–146. [Google Scholar] [CrossRef]
- EFSA. Opinion of the scientific panel on additives and products or substances used in animal feed on the safety and efficacy of the microbiological product “035”, a preparation of Bacillus subtilis, as a feed additive for chickens for fattening in accordance with Regulation (EC) No 1831/2003. EFSA J. 2006, 406, 1–11. [Google Scholar]
- Logan, N.A. Safety of Aerobic Endospore-forming Bacteria. In Bacterial Spore Formers; Ricca, E., Henriques, A.O., Cutting, S.M., Eds.; Horizon Biosciences: Norfolk, UK, 2004. [Google Scholar]
- Phelan, R.W.; Barret, M.; Cotter, P.D.; O’Connor, P.M.; Chen, R.; Morrissey, J.P.; Dobson, A.D.; O’Gara, F.; Barbosa, T.M. Subtilomycin: A new lantibiotic from Bacillus subtilis strain MMA7 isolated from the marine sponge Haliclona simulans. Mar. Drugs 2013, 11, 1878–1898. [Google Scholar] [CrossRef]
- Ugras, S.; Sezen, K.; Kati, H.; Demirbag, Z. Purification and characterization of the bacteriocin Thuricin Bn1 produced by Bacillus thuringiensis subsp. kurstaki Bn1 isolated from a hazelnut pest. J. Microbiol. Biotechnol. 2013, 23, 167–176. [Google Scholar] [CrossRef]
- Ruiz-Ponte, C.; Samain, J.F.; Sanchez, J.L.; Nicolas, J.L. The benefit of a Roseobacter species on the survival of scallop larvae. Mar. Biotechnol. 1999, 1, 52–59. [Google Scholar]
- Gardiner, G.; Stanton, C.; Lynch, P.; Collins, J.; Fitzgerald, G.; Ross, R. Evaluation of cheddar cheese as a food carrier for delivery of a probiotic strain to the gastrointestinal tract. J. Dairy Sci. 1999, 82, 1379–1387. [Google Scholar] [CrossRef]
- Dunne, C.; Murphy, L.; Flynn, S.; O’Mahony, L.; O’Halloran, S.; Feeney, M.; Morrissey, D.; Thornton, G.; Fitzgerald, G.; Daly, C. Probiotics: From myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie Van Leeuwenhoek 1999, 76, 279–292. [Google Scholar] [CrossRef]
- Barbosa, T.M.; Serra, C.R.; La Ragione, R.M.; Woodward, M.J.; Henriques, A.O. Screening for Bacillus isolates in the broiler gastrointestinal tract. Appl. Environ. Microbiol. 2005, 71, 968–978. [Google Scholar] [CrossRef]
- Trapecar, M.; Leouffre, T.; Faure, M.; Jensen, H.E.; Granum, P.E.; Cencic, A.; Hardy, S.P. The use of a porcine intestinal cell model system for evaluating the food safety risk of Bacillus cereus probiotics and the implications for assessing enterotoxigenicity. APMIS 2011, 119, 877–884. [Google Scholar] [CrossRef]
- Kim, K.M.; Kim, M.J.; Kim, D.H.; Park, Y.S.; Kang, J.S. Characterization of Bacillus polyfermenticus KJS-2 as a probiotic. J. Microbiol. Biotechnol. 2009, 19, 1013–1018. [Google Scholar]
- Leser, T.; Knarreborg, A.; Worm, J. Germination and outgrowth of Bacillus subtilis and Bacillus licheniformis spores in the gastrointestinal tract of pigs. J. Appl. Microbiol. 2008, 104, 1025–1033. [Google Scholar] [CrossRef]
- Cartman, S.T.; La Ragione, R.M.; Woodward, M.J. Bacterial Spore Formers as Probiotics for Poultry. In Food Science and Technology Bulletin: Functional Foods; Gibson, G.R., Ed.; International Food Information Service: Shinfield, UK, 2008; Volume 4, p. 102. [Google Scholar]
- Prieto, M.L.; O’Sullivan, L.; Tan, S.P.; McLoughlin, P.; Hughes, H.; O’Donovan, O.; Kent, R.M.; Cassidy, J.P.; Lawlor, P.G.; Gardiner, G.E. Evaluation of the efficacy and safety of a marine-derived Bacillus strain for use as an in-feed probiotic for newly weaned pigs. PLoS One 2014, 9, e88599. [Google Scholar]
- Duc, L.H.; Hong, H.A.; Cutting, S.M. Germination of the spore in the gastrointestinal tract provides a novel route for heterologous antigen delivery. Vaccine 2003, 21, 4215–4224. [Google Scholar]
- Hoa, N.T.; Baccigalupi, L.; Huxham, A.; Smertenko, A.; Van, P.H.; Ammendola, S.; Ricca, E.; Cutting, S.M. Characterization of Bacillus species used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders. Appl. Environ. Microbiol. 2000, 66, 5241–5247. [Google Scholar] [CrossRef]
- EFSA. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2012 update). EFSA J. 2012, 10, 3020. [Google Scholar]
- EFSA. Technical guidance on the assessment of the toxigenic potential of Bacillus species used in animal nutrition. EFSA J. 2011, 9, 2445. [Google Scholar]
- Patel, A.K.; Deshattiwar, M.K.; Chaudhari, B.L.; Chincholkar, S.B. Production, purification and chemical characterization of the catecholate siderophore from potent probiotic strains of Bacillus spp. Bioresour. Technol. 2009, 100, 368–373. [Google Scholar] [CrossRef]
- Gardiner, G.E.; Casey, P.G.; Casey, G.; Lynch, P.B.; Lawlor, P.G.; Hill, C.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P. Relative ability of orally administered Lactobacillus murinus to predominate and persist in the porcine gastrointestinal tract. Appl. Environ. Microbiol. 2004, 70, 1895–1906. [Google Scholar] [CrossRef]
- Nicholson, W.L.; Setlow, P. Sporulation, Germination and Outgrowth. In Molecular Biology Methods for Bacillus; Hardwood, C.R., Cutting, S.M., Eds.; Chichester: New York, NY, USA, 1990; p. 581. [Google Scholar]
- Casey, P.G.; Casey, G.D.; Gardiner, G.E.; Tangney, M.; Stanton, C.; Ross, R.P.; Hill, C.; Fitzgerald, G.F. Isolation and characterization of anti-Salmonella lactic acid bacteria from the porcine gastrointestinal tract. Lett. Appl. Microbiol. 2004, 39, 431–438. [Google Scholar] [CrossRef]
- Dobson, A.; Crispie, F.; Rea, M.C.; O’Sullivan, O.; Casey, P.G.; Lawlor, P.G.; Cotter, P.D.; Ross, P.; Gardiner, G.E.; Hill, C. Fate and efficacyof lacticin 3147-producing Lactococcus lactis in the mammalian gastrointestinal tract. FEMS Microbiol. Ecol. 2011, 76, 602–614. [Google Scholar] [CrossRef]
- Hsueh, Y.H.; Somers, E.B.; Lereclus, D.; Wong, A.C.L. Biofilm formation by Bacillus cereus is influenced by PlcR, a pleiotropic regulator. Appl. Environ. Microbiol. 2006, 72, 5089–5092. [Google Scholar] [CrossRef]
- Connelly, M.B.; Young, G.M.; Sloma, A. Extracellular proteolytic activity plays a central role in swarming motility in Bacillus subtilis. J. Bacteriol. 2004, 186, 4159–4167. [Google Scholar]
- Guinebretiere, M.H.; Broussolle, V.; Nguyen-The, C. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 2002, 40, 3053–3056. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals: Approved Standard; CLSI: Wayne, PA, USA, 2013; Volume 33. [Google Scholar]
- SAS/STAT. User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2010. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Prieto, M.L.; O'Sullivan, L.; Tan, S.P.; McLoughlin, P.; Hughes, H.; Gutierrez, M.; Lane, J.A.; Hickey, R.M.; Lawlor, P.G.; Gardiner, G.E. In Vitro Assessment of Marine Bacillus for Use as Livestock Probiotics. Mar. Drugs 2014, 12, 2422-2445. https://doi.org/10.3390/md12052422
Prieto ML, O'Sullivan L, Tan SP, McLoughlin P, Hughes H, Gutierrez M, Lane JA, Hickey RM, Lawlor PG, Gardiner GE. In Vitro Assessment of Marine Bacillus for Use as Livestock Probiotics. Marine Drugs. 2014; 12(5):2422-2445. https://doi.org/10.3390/md12052422
Chicago/Turabian StylePrieto, Maria Luz, Laurie O'Sullivan, Shiau Pin Tan, Peter McLoughlin, Helen Hughes, Montserrat Gutierrez, Jonathan A. Lane, Rita M. Hickey, Peadar G. Lawlor, and Gillian E. Gardiner. 2014. "In Vitro Assessment of Marine Bacillus for Use as Livestock Probiotics" Marine Drugs 12, no. 5: 2422-2445. https://doi.org/10.3390/md12052422
APA StylePrieto, M. L., O'Sullivan, L., Tan, S. P., McLoughlin, P., Hughes, H., Gutierrez, M., Lane, J. A., Hickey, R. M., Lawlor, P. G., & Gardiner, G. E. (2014). In Vitro Assessment of Marine Bacillus for Use as Livestock Probiotics. Marine Drugs, 12(5), 2422-2445. https://doi.org/10.3390/md12052422