Biosynthetic Functional Gene Analysis of Bis-Indole Metabolites from 25D7, a Clone Derived from a Deep-Sea Sediment Metagenomic Library
Abstract
:1. Introduction
2. Results and Discussion
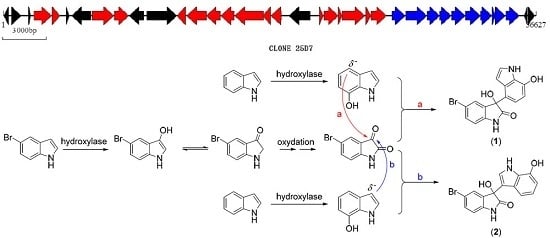
2.1. Bromo-Substituted Substrate Fermentation and LC-MS-Guided Separation
2.2. Structural Identification of the Two New Bromodiindoles
2.3. Biosynthetic Functional Gene Analysis of Bromo-Diindoles
2.4. Cytotoxic Activity of 5-Bromodiindoles
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fermentation
3.3. Extraction and Isolation
3.4. Transposon Mutagenesis and Sequence Analysis
3.5. Cytotoxic Activity
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Organ. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, V.; Øvreås, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Liu, Q.-Y.; Zhou, T.; Zhao, Y.-Y.; Chen, L.; Gong, M.-W.; Xia, Q.-W.; Ying, M.-G.; Zheng, Q.-H.; Zhang, Q.-Q. Antitumor Effects and Related Mechanisms of Penicitrinine A, a Novel Alkaloid with a Unique Spiro Skeleton from the Marine Fungus Penicillium citrinum. Mar. Drugs 2015, 13, 4733–4753. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Lu, X.; Ke, A.; Zheng, Z.; Lin, J.; Hao, W.; Zhu, J.; Fan, Y.; Ding, Y.; Jiang, Q. Three novel members of angucycline group from Streptomyces sp. N05WA963. J. Antibiot. 2011, 64, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.-Y.; Brady, S.F. Discovery of indolotryptoline antiproliferative agents by homology-guided metagenomic screening. Proc. Natl. Acad. Sci. USA 2013, 110, 2478–2483. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.S.; Brady, S.F. Arimetamycin A: Improving Clinically Relevant Families of Natural Products through Sequence-Guided Screening of Soil Metagenomes. Angew. Chem. Int. Ed. 2013, 52, 11063–11067. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.-Y.; Brady, S.F. Discovery and synthetic refactoring of tryptophan dimer gene clusters from the environment. J. Am. Chem. Soc. 2013, 135, 17906–17912. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tang, X.X.; Zheng, M.; Yi, Z.W.; Xiao, X.; Qiu, Y.K.; Wu, Z. A novel indole alkaloid from deep-sea sediment metagenomic clone-derived Escherichia coli fermentation broth. J. Asian Nat. Prod. Res. 2011, 13, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Tang, X.-X.; Chen, L.; Yi, Z.-W.; Fang, M.-J.; Wu, Z.; Qiu, Y.-K. Two New Cytotoxic Indole Alkaloids from a Deep-Sea Sediment. Derived Metagenomic Clone. Mar. Drugs 2014, 12, 2156–2163. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Kukita, A.; Akiyama, K.; Naito, T.; Uemura, D. Isolation and structure of a novel biindole pigment substituted with an ethyl group from a metagenomic library derived from the marine sponge Halichondria okadai. Chem. Lett. 2012, 41, 728–729. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, X.; Shao, Z.; Ren, J.; Liu, D.; Proksch, P.; Lin, W. Indole-Based alkaloids from deep-sea bacterium Shewanella piezotolerans with antitumor activities. J. Antibiot. 2014, 67, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.; Chimni, S.S. Asymmetric addition of indoles to isatins catalysed by bifunctional modified cinchona alkaloid catalysts. Chem. Eur. J. 2010, 16, 7709–7713. [Google Scholar] [CrossRef] [PubMed]
- Celik, A.; Speight, R.E.; Turner, N.J. Identification of broad specificity P450CAM variants by primary screening against indole as substrate. Chem. Commun. 2005, 7, 3652–3654. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P.; Sorrells, J.L.; Schmitt, S.; Krauser, J.A.; Aryal, P.; Meijer, L. Generation of New Protein Kinase Inhibitors Utilizing Cytochrome P450 Mutant Enzymes for Indigoid Synthesis. J. Med. Chem. 2004, 47, 3236–3241. [Google Scholar] [CrossRef] [PubMed]
- Berry, A.; Dodge, T.C.; Pepsin, M.; Weyler, W. Application of metabolic engineering to improve both the production and use of biotech indigo. J. Ind. Microbiol. Biotechnol. 2002, 28, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R. The metagenomics of soil. Nat. Rev. Microbiol. 2005, 3, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Menzella, H.G.; Chimni, S.S. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat. Biotechnol. 2005, 23, 1171–1176. [Google Scholar] [CrossRef] [PubMed]





| Position | δH (J in Hz) | δC, Multiple | 1H–1H COSY | HMBC | |
|---|---|---|---|---|---|
| 1-NH | 10.58 br.s | n.o. | |||
| 2 | 178.8 | C | |||
| 3 | 77.8 | C | |||
| 3a | 137.2 | C | |||
| 4 | 7.07 overlapped | 127.7 | CH | H-6 | C-3, 5, 6, 7a |
| 5 | 113.7 | C | |||
| 6 | 7.40 dd (8.3, 1.5) | 132 | CH | H-4, 7 | C-4, 5, 7a |
| 7 | 6.90 d (8.2) | 112.1 | CH | H-6 | C-3, 3a, 5 |
| 7a | 141.8 | C | |||
| 1′-NH | 10.95 s | H-2′, 3′ | C-2′, 3′, 3′a | ||
| 2′ | 7.07 overlapped | 124.8 | CH | H-1′, 3′ | C-3′, 3′a, 7′a |
| 3′ | 5.91 br.s | 101 | CH | H-2′ | C-2′, 3′a, 7′a |
| 3′a | 126.6 | C | |||
| 4′ | 122.7 | C | |||
| 5′ | 7.06 d (8.1) | 117.4 | CH | H-6′ | C-2, 3, 3′a, 6′, 7′ |
| 6′ | 6.51 d (8.1) | 104.8 | CH | H-5′ | C-4′, 7′, 7′a |
| 7′ | 144 | C | |||
| 7′a | 126.3 | C | |||
| 3-OH | 6.58 br.s | C-2, 3, 3a, 4′ | |||
| 7′-OH | 9.67 br.s | n.o. | |||
| Position | δH (J in Hz) | δC, Multiple | 1H–1H COSY | HMBC | |
|---|---|---|---|---|---|
| 1-NH | 10.49 s | n.o. | |||
| 2 | 178.4 | C | |||
| 3 | 75.4 | C | |||
| 3a | 136.5 | C | |||
| 4 | 7.27 br.d (2.0) | 127.8 | CH | H-6 | C-3, 5, 6, 7a |
| 5 | 113.7 | C | |||
| 6 | 7.42 dd (8.3, 2.0) | 132 | CH | H-4, 7 | C-4, 5, 7a |
| 7 | 6.87 d (8.3) | 112.2 | CH | H-6 | C-3, 3a, 5 |
| 7a | 141.4 | C | |||
| 1′-NH | 10.88 br. S | H-2′ | C-2′, 3′, 3′a | ||
| 2′ | 7.03 d (2.4) | 123.4 | CH | H-1′ | C-3, 3′, 3′a, 7′a |
| 3′ | 115.5 | C | |||
| 3′a | 126.9 | C | |||
| 4′ | 6.69 d (7.5) | 111.3 | CH | H-5′, 6′ | C-3′, 6′, 7′a |
| 5′ | 6.66 t (7.5) | 119.9 | CH | H-4′, 6′ | C-3′, 3′a, 6′, 7′ |
| 6′ | 6.45 d (7.3) | 105.9 | CH | H-4′, 5′ | C-4′, 7′, 7′a |
| 7′ | 144.1 | C | |||
| 7′a | 127.4 | C | |||
| 3-OH | 6.48 br.s | n.o. | |||
| 7′-OH | 9.57 br.s | n.o. | |||
| CDS No. | Predicted Gene | BLAST Result |
|---|---|---|
| 20 | phenol hydroxylase subunit | 83aa, 89% identity to Marinobacter algicola DG893 |
| 21 | phenol hydroxylase component phL | 333aa, 85% identity to Pseudomonas sp. OX1 |
| 22 | phenol hydroxylase component phM | 89aa, 96% identity to Pseudomonas sp. OX1 |
| 23 | phenol hydroxylase P3 protein | 515aa, 94% identity to M. algicola DG893 |
| 24 | phenol hydroxylase conserved region | 119aa, 84% identity to M. algicola DG893 |
| 25 | ferredoxin: oxidoreductase FAD/NAD(P)-binding | 353aa, 96% identity to M. algicola DG893 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, X.; Tang, X.-X.; Qin, D.; Yi, Z.-W.; Fang, M.-J.; Wu, Z.; Qiu, Y.-K. Biosynthetic Functional Gene Analysis of Bis-Indole Metabolites from 25D7, a Clone Derived from a Deep-Sea Sediment Metagenomic Library. Mar. Drugs 2016, 14, 107. https://doi.org/10.3390/md14060107
Yan X, Tang X-X, Qin D, Yi Z-W, Fang M-J, Wu Z, Qiu Y-K. Biosynthetic Functional Gene Analysis of Bis-Indole Metabolites from 25D7, a Clone Derived from a Deep-Sea Sediment Metagenomic Library. Marine Drugs. 2016; 14(6):107. https://doi.org/10.3390/md14060107
Chicago/Turabian StyleYan, Xia, Xi-Xiang Tang, Dan Qin, Zhi-Wei Yi, Mei-Juan Fang, Zhen Wu, and Ying-Kun Qiu. 2016. "Biosynthetic Functional Gene Analysis of Bis-Indole Metabolites from 25D7, a Clone Derived from a Deep-Sea Sediment Metagenomic Library" Marine Drugs 14, no. 6: 107. https://doi.org/10.3390/md14060107
APA StyleYan, X., Tang, X.-X., Qin, D., Yi, Z.-W., Fang, M.-J., Wu, Z., & Qiu, Y.-K. (2016). Biosynthetic Functional Gene Analysis of Bis-Indole Metabolites from 25D7, a Clone Derived from a Deep-Sea Sediment Metagenomic Library. Marine Drugs, 14(6), 107. https://doi.org/10.3390/md14060107