Identification of Angiotensin I-Converting Enzyme Inhibitory Peptides Derived from Enzymatic Hydrolysates of Razor Clam Sinonovacula constricta
Abstract
:1. Introduction
2. Results and Discussion
2.1. Production of Enzymatic Hydrolysates
2.1.1. Proximate Composition of Razor Clam
2.1.2. Degree of Hydrolysis and ACE-Inhibitory Activity of Hydrolysates by Different Proteases
2.1.3. Peptide Content and ACE-Inhibitory Activity of Ultra-Filtration Fractions
2.2. Identification of ACE-Inhibitory Peptides
2.2.1. Isolation and Purification of ACE-Inhibitory Peptides
2.2.2. Determination of Amino Acids Sequence
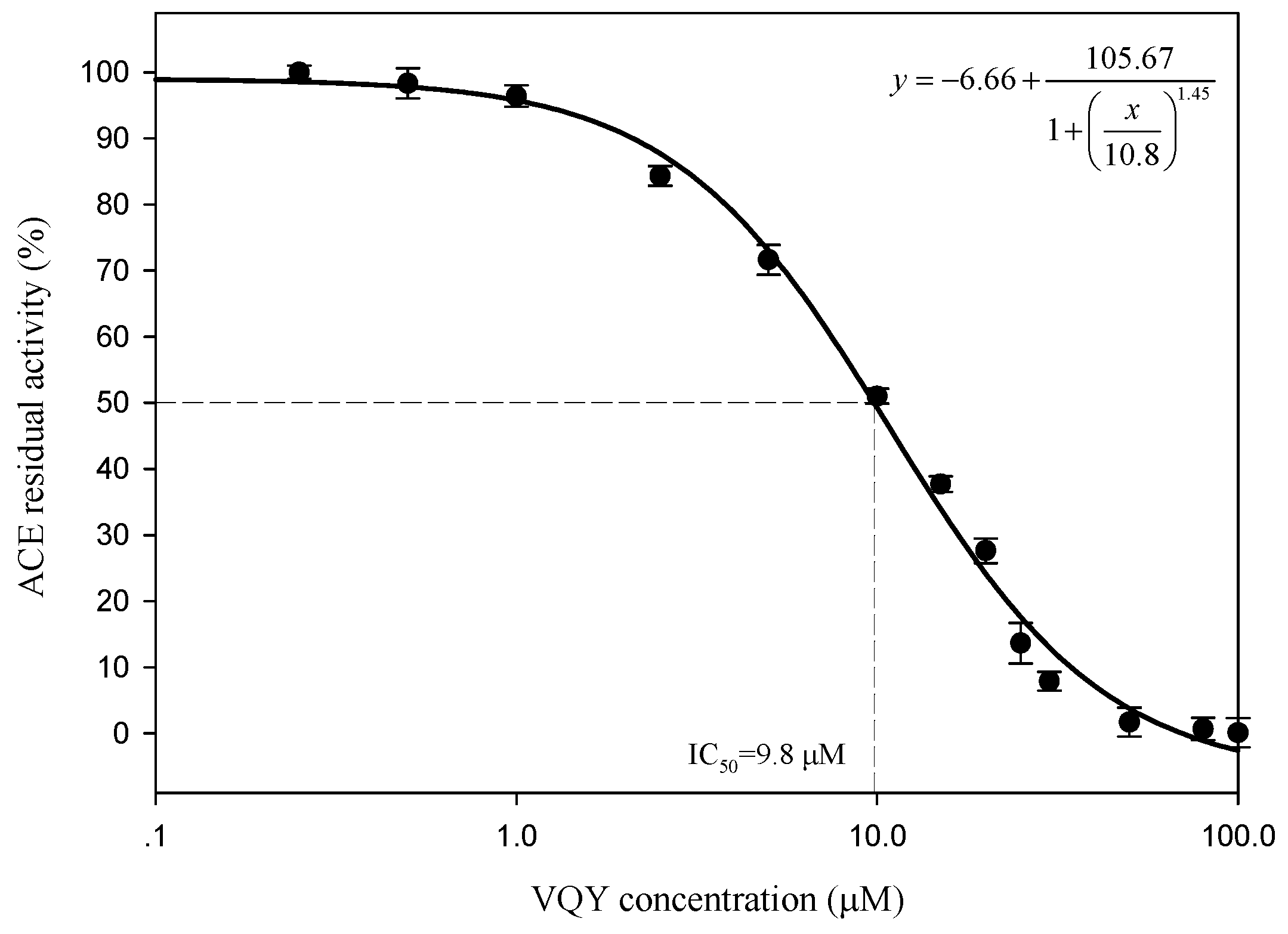
2.2.3. IC50 Value and Inhibition Pattern of Val-Gln-Tyr
3. Materials and Methods
3.1. Materials
3.2. Preparation of Crude Proteases from Actinomucor elegans T3
3.3. Enzymatic Hydrolysis
3.4. Analytical Methods
3.4.1. Angiotensin-Converting Enzyme Inhibition Assay
3.4.2. Degree of Hydrolysis Evaluation
3.4.3. Determination of Peptide Content
3.5. Purification and Identification of ACE-Inhibitory Peptides
3.5.1. Gel Filtration Chromatography
3.5.2. Reversed-Phase High-Performance Liquid Chromatography
3.5.3. Identification of the Amino Acid Sequence by MALDI/TOF-TOF MS/MS
3.5.4. Determination of ACE-Inhibition Pattern
3.6. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Poulter, N.R.; Prabhakaran, D.; Caulfield, M. Hypertension. Lancet 2015, 386, 730–731. [Google Scholar] [CrossRef]
- Lackland, D.T.; Weber, M.A. Global burden of cardiovascular disease and stroke: Hypertension at the core. Can. J. Cardiol. 2015, 31, 569–571. [Google Scholar] [CrossRef] [PubMed]
- George, B.; Pantelis, S.; Rajiv, A.; Luis, R. Review of blood pressure control rates and outcomes. J. Am. Soc. Hypertens. 2014, 8, 127–141. [Google Scholar]
- FerrãO, F.M.; Lara, L.S.; Lowe, J. Renin-angiotensin system in the kidney: What is new? World J. Nephrol. 2014, 3, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Cheung, R.C.; Ng, T.B.; Wong, J.H. Marine peptides: Bioactivities and applications. Mar. Drugs 2015, 13, 4006–4043. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wu, C.; Liu, D.; Yang, X.; Huang, J.; Zhang, J.; Liao, B.; He, H.; Li, H. Overview of antioxidant peptides derived from marine resources: The sources, characteristic, purification, and evaluation methods. Appl. Biochem. Biotechnol. 2015, 176, 1815–1833. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Bai, L.; Zhu, L.; Yang, L.; Zhang, X. Marine algae-derived bioactive peptides for human nutrition and health. J. Agric. Food Chem. 2014, 62, 9211–9222. [Google Scholar] [CrossRef] [PubMed]
- Bartneck, M.; Heffels, K.H.; Pan, Y.; Bovi, M.; Zwadlo-Klarwasser, G.; Groll, J. Inducing healing-like human primary macrophage phenotypes by 3D hydrogel coated nanofibres. Biomaterials 2012, 33, 4136–4146. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.H.; Bartelt, D.C.; Greene, L.J. Isolation of bradykinin-potentiating peptides from Bothrops jararaca venom. Biochemistry 1970, 9, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- Saadi, S.; Saari, N.; Anwar, F.; Abdul Hamid, A.; Ghazali, H.M. Recent advances in food biopeptides: Production, biological functionalities and therapeutic applications. Biotechnol. Adv. 2015, 33, 80–116. [Google Scholar] [CrossRef] [PubMed]
- Shiozaki, K.; Shiozaki, M.; Masuda, J.; Yamauchi, A.; Ohwada, S.; Nakano, T.; Yamaguchi, T.; Saito, T.; Muramoto, K.; Sato, M. Identification of oyster-derived hypotensive peptide acting as angiotensin-I-converting enzyme inhibitor. Fish. Sci. 2010, 76, 865–872. [Google Scholar] [CrossRef]
- Wang, J.; Hu, J.; Cui, J.; Bai, X.; Du, Y.; Miyaguchi, Y.; Lin, B. Purification and identification of a ACE inhibitory peptide from oyster proteins hydrolysate and the antihypertensive effect of hydrolysate in spontaneously hypertensive rats. Food Chem. 2008, 111, 302–308. [Google Scholar] [CrossRef] [PubMed]
- He, H.L.; Chen, X.L.; Sun, C.Y.; Zhang, Y.Z.; Zhou, B.C. Analysis of novel angiotensin-I-converting enzyme inhibitory peptides from protease-hydrolyzed marine shrimp Acetes chinensis. J. Pept. Sci. 2006, 12, 726–733. [Google Scholar]
- Tsai, J.S.; Pan, C.B.S. ACE-inhibitory peptides identified from the muscle protein hydrolysate of hard clam (Meretrix lusoria). Process Biochem. 2008, 43, 743–747. [Google Scholar] [CrossRef]
- Balti, R.; Nedjar-Arroume, N.; Adje, E.Y.; Guillochon, D.; Nasri, M. Analysis of novel angiotensin I-converting enzyme inhibitory peptides from enzymatic hydrolysates of cuttlefish (Sepia officinalis) muscle proteins. J. Agric. Food Chem. 2010, 58, 3840–3846. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Dong, L.; Niu, D.; Meng, S.; Zhang, B.; Liu, D.; Hu, S.; Li, J. Identification of immune genes of the Agamaki clam (Sinonovacula constricta) by sequencing and bioinformatic analysis of ests. Mar. Biotechnol. 2010, 12, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Karnjanapratum, S.; Benjakul, S.; Kishimura, H.; Tsai, Y.H. Chemical compositions and nutritional value of Asian hard clam (Meretrix lusoria) from the coast of Andaman Sea. Food Chem. 2013, 141, 4138–4145. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; An, Y.K.; Choi, S.D.; Kim, J. Proximate composition in the muscle and viscera of five Veneridae clams (bivalvia) from southern coast of Korea. Korean J. Matacol. 2008, 24, 67–72. [Google Scholar]
- Laxmilatha, P. Proximate composition of the surf clam Mactra violacea (Gmelin 1791). Indian J. Fish. 2009, 56, 147–150. [Google Scholar]
- Bougatef, A.; Nedjar-Arroume, N.; Ravallec-Plé, R.; Leroy, Y.; Guillochon, D.; Barkia, A.; Nasri, M. Angiotensin I-converting enzyme (ACE) inhibitory activities of sardinelle (Sardinella aurita) by-products protein hydrolysates obtained by treatment with microbial and visceral fish serine proteases. Food Chem. 2008, 111, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Giménez, B.; Alemán, A.; Montero, P.; Gómez-Guillén, M. Antioxidant and functional properties of gelatin hydrolysates obtained from skin of sole and squid. Food Chem. 2009, 114, 976–983. [Google Scholar] [CrossRef]
- Klompong, V.; Benjakul, S.; Kantachote, D.; Shahidi, F. Antioxidative activity and functional properties of protein hydrolysate of yellow stripe trevally (Selaroides leptolepis) as influenced by the degree of hydrolysis and enzyme type. Food Chem. 2007, 102, 1317–1327. [Google Scholar] [CrossRef]
- Mahmoodani, F.; Ghassem, M.; Babji, A.S.; Yusop, S.M.; Khosrokhavar, R. ACE inhibitory activity of pangasius catfish (Pangasius sutchi) skin and bone gelatin hydrolysate. J. Food Sci. Technol. 2014, 51, 1847–1856. [Google Scholar] [CrossRef] [PubMed]
- Benjakul, S.; Morrissey, M.T. Protein hydrolysates from pacific whiting solid wastes. J. Agric. Food Chem. 1997, 45, 3423–3430. [Google Scholar] [CrossRef]
- Nguyen, H.T.M.; Sylla, K.S.B.; Randriamahatody, Z.; Donnay-Moreno, C.; Moreau, J.; Tran, L.T.; Bergé, J.P. Enzymatic hydrolysis of yellowfin tuna (Thunnus albacares) by-products using protamex protease. Food Technol. Biotechnol. 2011, 49, 48–55. [Google Scholar]
- Guerard, F.; Guimas, L.; Binet, A. Production of tuna waste hydrolysates by a commercial neutral protease preparation. J. Mol. Catal. B Enzym. 2002, 19, 489–498. [Google Scholar] [CrossRef]
- Hausch, F.; Shan, L.; Santiago, N.A.; Gray, G.M.; Khosla, C. Intestinal digestive resistance of immunodominant gliadin peptides. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 283, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Bamdad, F.; Wu, J.P.; Chen, L.Y. Effects of enzymatic hydrolysis on molecular structure and antioxidant activity of barley hordein. J. Cereal Sci. 2011, 54, 20–28. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Limited enzymic degradation of proteins: A new approach in the industrial application of hydrolases. J. Chem. Technol. Biotechnol. 1982, 32, 138–156. [Google Scholar] [CrossRef]
- Wu, J.P.; Aluko, R.E.; Muir, A.D. Production of angiotensin I-converting enzyme inhibitory peptides from defatted canola meal. Bioresour. Technol. 2009, 100, 5283–5287. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M.; Forghani, B.; Ebrahimpour, A.; Abdul-Hamid, A.; Anwar, F.; Saari, N. In vitro and in vivo antihypertensive activity of palm kernel cake protein hydrolysates: Sequencing and characterization of potent bioactive peptides. Ind. Crop. Prod. 2015, 76, 112–120. [Google Scholar] [CrossRef]
- Zhang, Y.; Olsen, K.; Grossi, A.; Otte, J. Effect of pretreatment on enzymatic hydrolysis of bovine collagen and formation of ACE-inhibitory peptides. Food Chem. 2013, 141, 2343–2354. [Google Scholar] [CrossRef] [PubMed]
- Cheung, I.W.Y.; Li-Chan, E.C.Y. Application of taste sensing system for characterisation of enzymatic hydrolysates from shrimp processing by-products. Food Chem. 2014, 145, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Aissaoui, N.; Abidi, F.; Marzouki, M.N. ACE inhibitory and antioxidant activities of red scorpionfish (Scorpaena notata) protein hydrolysates. J. Food Sci. Technol. Mysore 2015, 52, 7092–7102. [Google Scholar] [CrossRef]
- Hernandez-Ledesma, B.; Contreras, M.D.; Recio, I. Antihypertensive peptides: Production, bioavailability and incorporation into foods. Adv. Colloid Interface Sci. 2011, 165, 23–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramanathan, N.; Schwager, S.L.U.; Sturrock, E.D.; Acharya, K.R. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature 2003, 421, 551–554. [Google Scholar]
- Tsai, J.S.; Lin, T.C.; Chen, J.L.; Pan, B.S. The inhibitory effects of freshwater clam (Corbicula fluminea, Muller) muscle protein hydrolysates on angiotensin I converting enzyme. Process Biochem. 2006, 41, 2276–2281. [Google Scholar] [CrossRef]
- Cristina, M.; Maria, D.M.Y.; Justo, P.; Hassan, L.; Julio, G.C.; Manuel, A.; Francisco, M.; Javier, V. Purification of an ACE inhibitory peptide after hydrolysis of sunflower (Helianthus annuus L.) protein isolates. J. Agric. Food Chem. 2004, 52, 1928–1932. [Google Scholar]
- Wu, J.; Ding, X. Hypotensive and physiological effect of angiotensin converting enzyme inhibitory peptides derived from soy protein on spontaneously hypertensive rats. J. Agric. Food Chem. 2001, 49, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Aluko, R.E.; Shuryo, N. Structural requirements of angiotensin I-converting enzyme inhibitory peptides: Quantitative structure-activity relationship study of di- and tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Rohrbach, M.S.; Williams, E.B.; Rolstad, R.A. Purification and substrate specificity of bovine angiotensin-converting enzyme. J. Biol. Chem. 1981, 256, 225–230. [Google Scholar] [PubMed]
- Helrich, K. Official Methods of Analysis of the AOAC, 14th ed.; Association of Official Analytical Chemists: Washington, DC, USA, 1990. [Google Scholar]
- Cushman, D.W.; Cheung, H.S. Spectrophotometric assay and properties of the angiotensin I-converting enzyme of rabbit lung. Biochem. Pharmacol. 1971, 20, 1637–1648. [Google Scholar] [CrossRef]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved method for determining food protein degree of hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Rao, S.Q.; Ju, T.; Sun, J.; Su, Y.J.; Xu, R.R.; Yang, Y.J. Purification and characterization of angiotensin I-converting enzyme inhibitory peptides from enzymatic hydrolysate of hen egg white lysozyme. Food Res. Int. 2012, 46, 127–134. [Google Scholar] [CrossRef]






| Composition | Contents (g/100 g Fresh Weight) |
|---|---|
| Moisture | 80.32 ± 0.53 |
| Protein | 13.68 ± 0.62 |
| Fat | 1.89 ± 0.13 |
| Carbohydrate | 2.13 ± 0.31 |
| Ash | 1.93 ± 0.08 |
| Protease | Source | Temperature (°C) | pH |
|---|---|---|---|
| Pepsin | porcine gastric mucosa | 37 | 2.0 |
| Trypsin | bovine pancreas | 37 | 8.0 |
| Alcalase | Bacillus licheniformis | 40 | 8.0 |
| Flavourzyme | Aspergillus oryzae | 50 | 6.0 |
| Crude proteases | Actinomucor elegans | 55 | 6.0 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Sadiq, F.A.; Fu, L.; Zhu, H.; Zhong, M.; Sohail, M. Identification of Angiotensin I-Converting Enzyme Inhibitory Peptides Derived from Enzymatic Hydrolysates of Razor Clam Sinonovacula constricta. Mar. Drugs 2016, 14, 110. https://doi.org/10.3390/md14060110
Li Y, Sadiq FA, Fu L, Zhu H, Zhong M, Sohail M. Identification of Angiotensin I-Converting Enzyme Inhibitory Peptides Derived from Enzymatic Hydrolysates of Razor Clam Sinonovacula constricta. Marine Drugs. 2016; 14(6):110. https://doi.org/10.3390/md14060110
Chicago/Turabian StyleLi, Yun, Faizan A. Sadiq, Li Fu, Hui Zhu, Minghua Zhong, and Muhammad Sohail. 2016. "Identification of Angiotensin I-Converting Enzyme Inhibitory Peptides Derived from Enzymatic Hydrolysates of Razor Clam Sinonovacula constricta" Marine Drugs 14, no. 6: 110. https://doi.org/10.3390/md14060110





