Structural Diversity, Biological Properties and Applications of Natural Products from Cyanobacteria. A Review †
Abstract
:1. Introduction
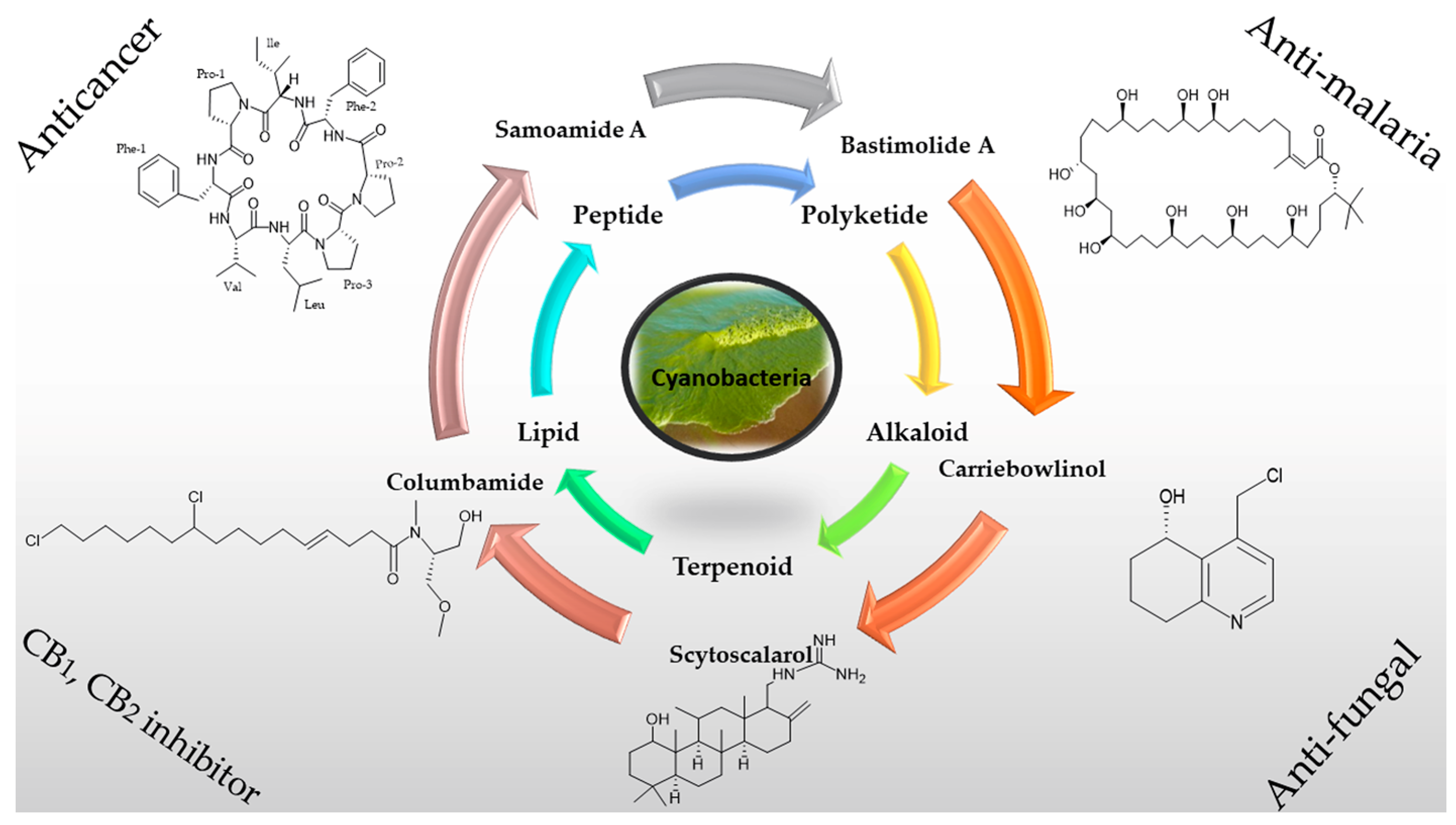
2. Chemical Diversity of Secondary Metabolites from Cyanobacteria
2.1. Peptides
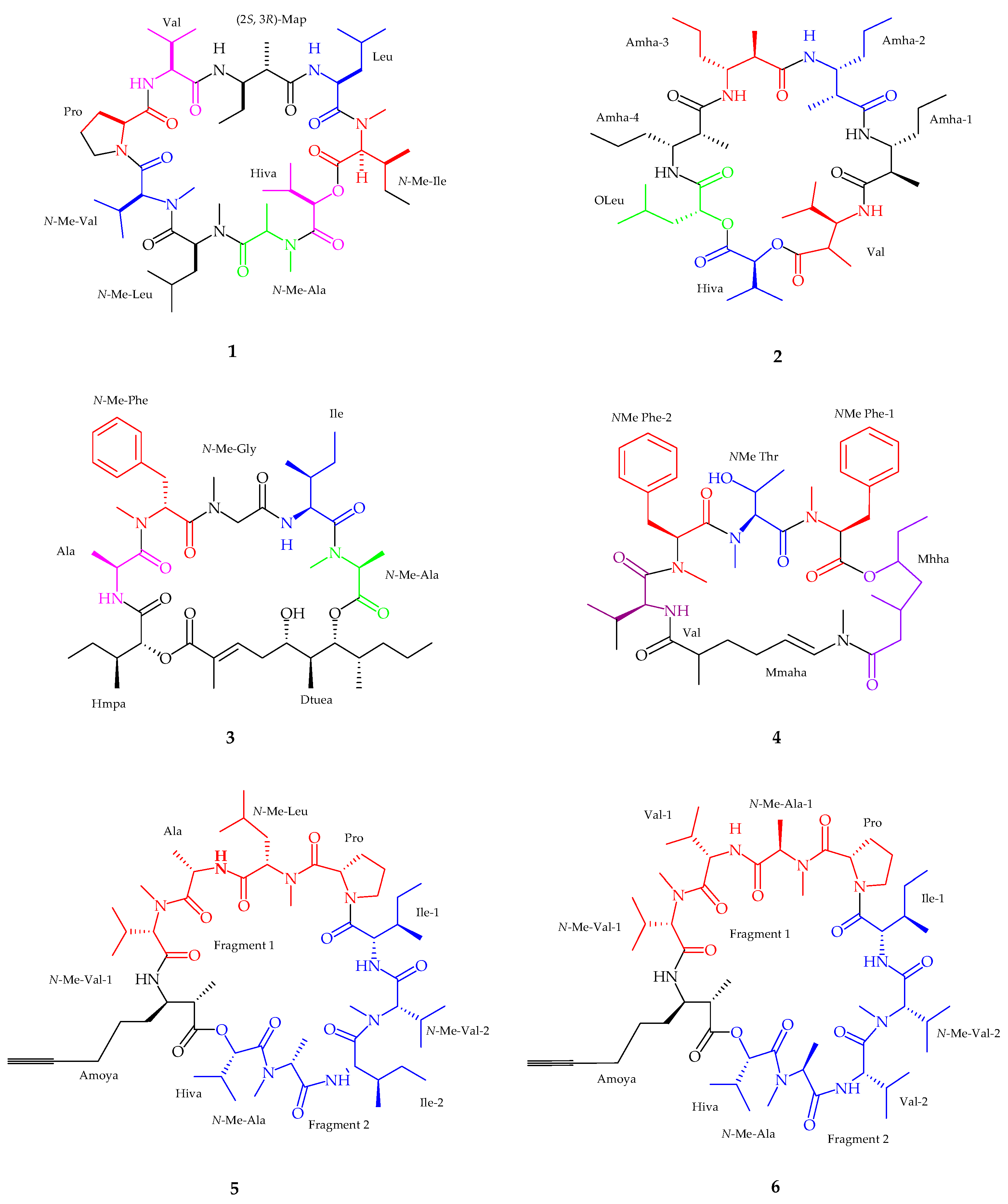
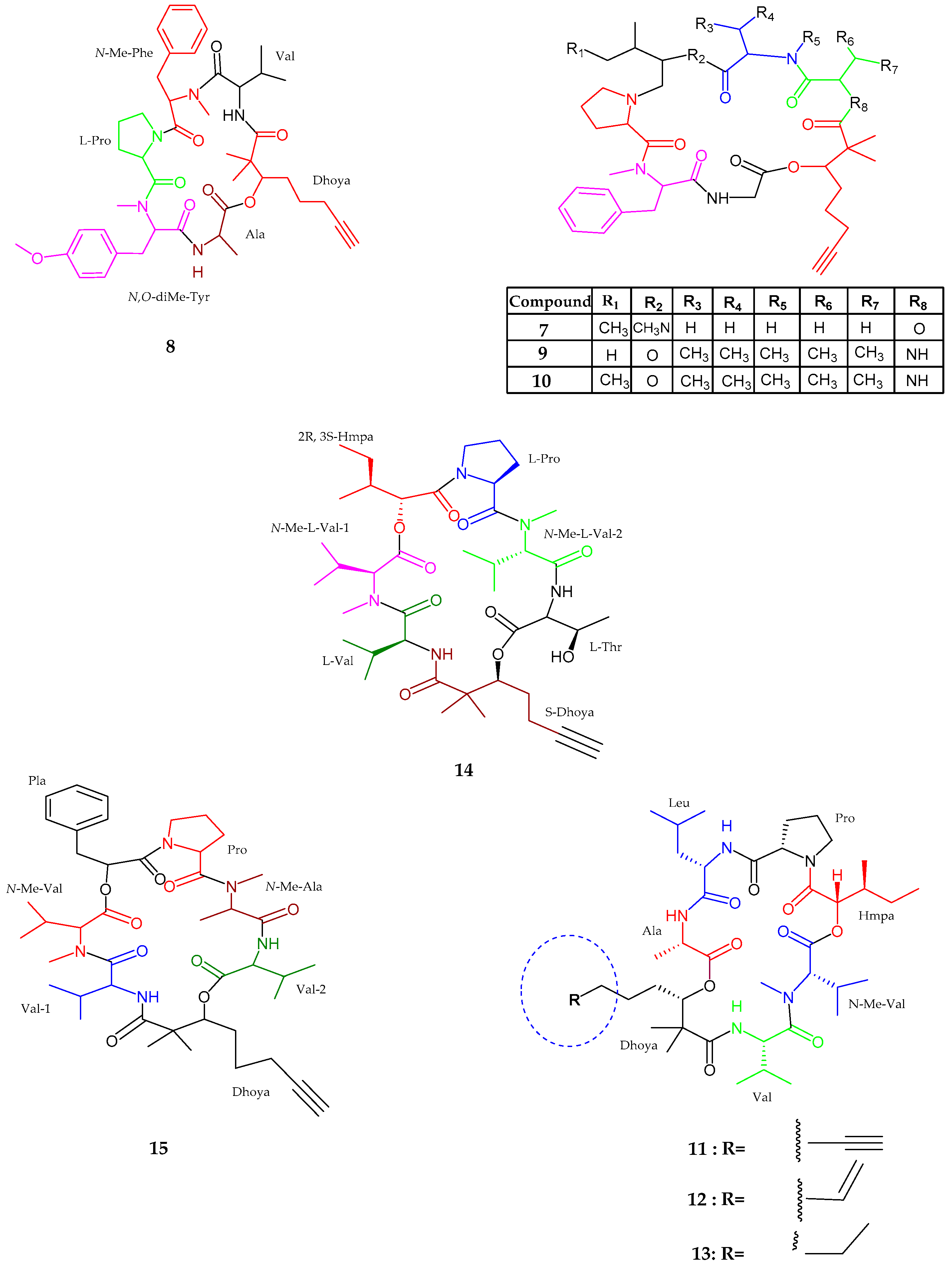
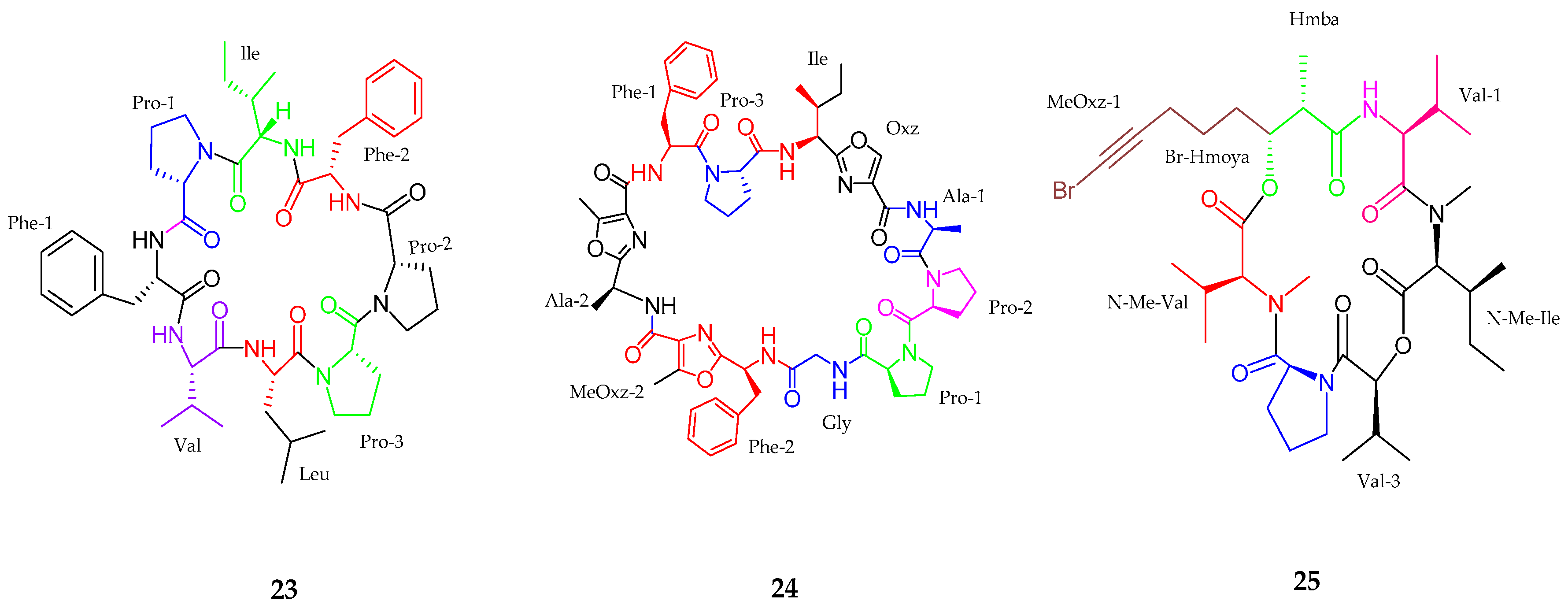
2.1.1. Cyclic Peptide
2.1.2. Linear Peptides
2.2. Polyketides
2.3. Alkaloids
2.4. Lipids
3. Applications
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef] [PubMed]
- Zarshenas, M.M.; Dabaghian, F.; Moein, M. An Overview on Phytochemical and Pharmacological Aspects of Echium amoenum. Nat. Prod. J. 2016, 6, 285–291. [Google Scholar] [CrossRef]
- Singh, P.; Rawat, M.S.M. Phytochemistry and Biological Activity Perspectives of Rheum Species. Nat. Prod. J. 2016, 6, 84–93. [Google Scholar] [CrossRef]
- Do Rosário Martins, M.; Costa, M. Marine cyanobacteria compounds with anticancer properties: Implication of apoptosis. In Handbook of Anticancer Drugs from Marine Origin; Springer: Berlin, Germany, 2015; pp. 621–647. [Google Scholar]
- Sharma, A.K.; Beniwal, V.; Kumar, R.; Thakur, K.; Sharma, R. Secondary Metabolites Derived from Actinomycetes: Iron Modulation and Their Therapeutic Potential. Nat. Prod. J. 2015, 5, 72–81. [Google Scholar] [CrossRef]
- Cheung, R.C.F.; Ng, T.B.; Wong, J.H. Marine peptides: Bioactivities and applications. Mar. Drugs 2015, 13, 4006–4043. [Google Scholar] [CrossRef] [PubMed]
- Jo, C.; Khan, F.F.; Khan, M.I.; Iqbal, J. Marine bioactive peptides: Types, structures, and physiological functions. Food Rev. Int. 2017, 33, 44–61. [Google Scholar] [CrossRef]
- Mazard, S.; Penesyan, A.; Ostrowski, M.; Paulsen, I.T.; Egan, S. Tiny microbes with a big impact: The role of cyanobacteria and their metabolites in shaping our future. Mar. Drugs 2016, 14, 97. [Google Scholar] [CrossRef] [PubMed]
- Rangnekar, S.S.; Khan, T. Novel Anti-Inflammatory Drugs from Marine Microbes. Nat. Prod. J. 2015, 5, 206–218. [Google Scholar] [CrossRef]
- Dixit, R.B.; Suseela, M.R. Cyanobacteria: Potential candidates for drug discovery. Antonie Van Leeuwenhoek 2013, 103, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, D.; Finking, R.; Marahiel, M.A. Nonribosomal peptides: From genes to products. Nat. Prod. Rep. 2003, 20, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Foster, R.A.; Kuypers, M.M.M.; Vagner, T.; Paerl, R.W.; Musat, N.; Zehr, J.P. Nitrogen fixation and transfer in open ocean diatom–cyanobacterial symbioses. ISME J. 2011, 5, 1484–1493. [Google Scholar] [CrossRef] [PubMed]
- Anjum, K.; Abbas, S.Q.; Akhter, N.; Shagufta, B.I.; Shah, S.A.A. Emerging biopharmaceuticals from bioactive peptides derived from marine organisms. Chem. Biol. Drug Des. 2017, 90, 12–30. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.T. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry 2007, 68, 954–979. [Google Scholar] [CrossRef] [PubMed]
- Gademann, K.; Portmann, C. Secondary metabolites from cyanobacteria: Complex structures and powerful bioactivities. Curr. Org. Chem. 2008, 12, 326–341. [Google Scholar] [CrossRef]
- Malathi, T.; Babu, M.R.; Mounika, T.; Snehalatha, D.; Rao, B.D. Screening of cyanobacterial strains for antibacterial activity. Phykos 2014, 44, 6–11. [Google Scholar]
- Shaieb, F.A.; Issa, A.A.-S.; Meragaa, A. Antimicrobial activity of crude extracts of cyanobacteria Nostoc commune and Spirulina platensis. Arch. Biomed. Sci. 2014, 2, 34–41. [Google Scholar]
- Mukund, S.; Sivasubramanian, V. Anticancer Activity of Oscillatoria Terebriformis Cyanobacteria in Human Lung Cancer Cell Line A549. Int. J. Appl. Biol. Pharm. Technol. 2014, 5, 34–45. [Google Scholar]
- El Semary, N.A.; Fouda, M. Anticancer activity of Cyanothece sp. strain extracts from Egypt: First record. Asian Pac. J. Trop. Biomed. 2015, 5, 992–995. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Menakha, M. Pharmaceutical applications of cyanobacteria—A review. J. Acute Med. 2015, 5, 15–23. [Google Scholar] [CrossRef]
- Dittmann, E.; Neilan, B.; Börner, T. Molecular biology of peptide and polyketide biosynthesis in cyanobacteria. Appl. Microbiol. Biotechnol. 2001, 57, 467–473. [Google Scholar] [PubMed]
- Volk, R.-B. Screening of microalgae for species excreting norharmane, a manifold biologically active indole alkaloid. Microbiol. Res. 2008, 163, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Sato, S.; Kotani, H.; Tanaka, A.; Asamizu, E.; Nakamura, Y.; Miyajima, N.; Hirosawa, M.; Sugiura, M.; Sasamoto, S.; et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential proteincoding regions. DNA Res. 1996, 3, 109–136. [Google Scholar] [CrossRef] [PubMed]
- Luan, G.; Qi, Y.; Wang, M.; Li, Z.; Duan, Y.; Tan, X.; Lu, X. Combinatory strategy for characterizing and understanding the ethanol synthesis pathway in cyanobacteria cell factories. Biotechnol. Biofuels 2015, 8, 184. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.D.; Fewer, D.P.; Talla, E.; Calteau, A.; Cai, F.; Tandeau de Marsac, N.; Rippka, R.; et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Chung-Davidson, Y.-W.; Bussy, U.; Li, W. Recent Advances and Applications of Experimental Technologies in Marine Natural Product Research. Mar. Drugs 2015, 13, 2694–2713. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, S.; Manogar, P.; Prabhu, S. Potential therapeutic targets and the role of technology in developing novel cannabinoid drugs from cyanobacteria. Biomed. Pharmacother. 2016, 83, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Zhang, J.; He, S.; Yan, X. New Peptides Isolated from Marine Cyanobacteria, an Overview over the Past Decade. Mar. Drugs 2017, 15, 132. [Google Scholar] [CrossRef] [PubMed]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Parihar, P.; Singh, M.; Bajguz, A.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Uncovering Potential Applications of Cyanobacteria and Algal Metabolites in Biology, Agriculture and Medicine: Current Status and Future Prospects. Front. Microbiol. 2017, 8, 515. [Google Scholar] [CrossRef] [PubMed]
- Minich, S.S. Brentuximab vedotin: A new age in the treatment of Hodgkin lymphoma and anaplastic large cell lymphoma. Ann. Pharmacother. 2012, 46, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, Y.; Iwasaki, A.; Sumimoto, S.; Suenaga, K. Urumamide, a novel chymotrypsin inhibitor with a β-amino acid from a marine cyanobacterium Okeania sp. Tetrahedron Lett. 2016, 57, 4213–4216. [Google Scholar] [CrossRef]
- Fenner, A.M.; Engene, N.; Spadafora, C.; Gerwick, W.H.; Balunas, M.J. Medusamide A, a Panamanian cyanobacterial depsipeptide with multiple β-Amino acids. Org. Lett. 2016, 18, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Sueyoshi, K.; Kaneda, M.; Sumimoto, S.; Oishi, S.; Fujii, N.; Suenaga, K.; Teruya, T. Odoamide, a cytotoxic cyclodepsipeptide from the marine cyanobacterium Okeania sp. Tetrahedron 2016, 72, 5472–5478. [Google Scholar] [CrossRef]
- Tan, L.T.; Okino, T.; Gerwick, W.H. Bouillonamide: A mixed polyketide–peptide cytotoxin from the marine cyanobacterium Moorea bouillonii. Mar. Drugs 2013, 11, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Vining, O.B.; Medina, R.A.; Mitchell, E.A.; Videau, P.; Li, D.; Serrill, J.D.; Kelly, J.X.; Gerwick, W.H.; Proteau, P.J.; Ishmael, J.E. Depsipeptide companeramides from a panamanian marine cyanobacterium associated with the coibamide producer. J. Nat. Prod. 2015, 78, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Almaliti, J.; Malloy, K.L.; Glukhov, E.; Spadafora, C.; Gutiérrez, M.; Gerwick, W.H. Dudawalamides A–D, Antiparasitic Cyclic Depsipeptides from the Marine Cyanobacterium Moorea producens. J. Nat. Prod. 2017, 80, 1827–1836. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Shiota, I.; Sumimoto, S.; Matsubara, T.; Sato, T.; Suenaga, K. Kohamamides A, B, and C, Cyclic Depsipeptides from an Okeania sp. Marine Cyanobacterium. J. Nat. Prod. 2017, 80, 1948–1952. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, P.D.; Byrum, T.; Liu, W.T.; Dorrestein, P.C.; Gerwick, W.H. Viequeamide A, a cytotoxic member of the kulolide superfamily of cyclic depsipeptides from a marine button cyanobacterium. J. Nat. Prod. 2012, 75, 1560–1570. [Google Scholar] [CrossRef] [PubMed]
- Montaser, R.; Abboud, K.A.; Paul, V.J.; Luesch, H. Pitiprolamide, a proline-rich dolastatin 16 analogue from the marine cyanobacterium Lyngbya majuscula from guam. J. Nat. Prod. 2011, 74, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Montaser, R.; Paul, V.J.; Luesch, H. Pitipeptolides C–F, antimycobacterial cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula from Guam. Phytochemistry 2011, 72, 2068–2074. [Google Scholar] [CrossRef] [PubMed]
- Thornburg, C.C.; Cowley, E.S.; Sikorska, J.; Shaala, L.A.; Ishmael, J.E.; Youssef, D.T.A.; McPhail, K.L. Apratoxin H and apratoxin A sulfoxide from the red sea cyanobacterium Moorea producens. J. Nat. Prod. 2013, 76, 1781–1788. [Google Scholar] [CrossRef] [PubMed]
- Naman, C.B.; Rattan, R.; Nikoulina, S.E.; Lee, J.; Miller, B.W.; Moss, N.A.; Armstrong, L.; Boudreau, P.D.; Debonsi, H.M.; Valeriote, F.A.; et al. Integrating Molecular Networking and Biological Assays To Target the Isolation of a Cytotoxic Cyclic Octapeptide, Samoamide A, from an American Samoan Marine Cyanobacterium. J. Nat. Prod. 2017, 80, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.A.V.; Al-Lihaibi, S.S.; Alarif, W.M.; Abdel-Lateff, A.; Nogata, Y.; Washio, K.; Morikawa, M.; Okino, T. Wewakazole B, a Cytotoxic Cyanobactin from the Cyanobacterium Moorea producens Collected in the Red Sea. J. Nat. Prod. 2016, 79, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Nogle, L.M.; Marquez, B.L.; Gerwick, W.H. Wewakazole, a novel cyclic dodecapeptide from a papua new guinea Lyngbya majuscula. Org. Lett. 2003, 5, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Sueyoshi, K.; Kudo, T.; Yamano, A.; Sumimoto, S.; Iwasaki, A.; Suenaga, K.; Teruya, T. Odobromoamide, a terminal alkynyl bromide-containing cyclodepsipeptide from the marine cyanobacterium Okeania sp. Bull. Chem. Soc. Jpn. 2017, 90, 436–440. [Google Scholar] [CrossRef]
- Spoof, L.; Błaszczyk, A.; Meriluoto, J.; Cegłowska, M.; Mazur-Marzec, H. Structures and activity of new anabaenopeptins produced by Baltic Sea cyanobacteria. Mar. Drugs 2016, 14, 8. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Ohno, O.; Sumimoto, S.; Suda, S.; Suenaga, K. Maedamide, a novel chymotrypsin inhibitor from a marine cyanobacterial assemblage of Lyngbya sp. Tetrahedron Lett. 2014, 55, 4126–4128. [Google Scholar] [CrossRef]
- Gunasekera, S.P.; Imperial, L.; Garst, C.; Ratnayake, R.; Dang, L.H.; Paul, V.J.; Luesch, H. Caldoramide, a Modified Pentapeptide from the Marine Cyanobacterium Caldora penicillata. J. Nat. Prod. 2016, 79, 1867–1871. [Google Scholar] [CrossRef] [PubMed]
- Mevers, E.; Haeckl, F.P.J.; Boudreau, P.D.; Byrum, T.; Dorrestein, P.C.; Valeriote, F.A.; Gerwick, W.H. Lipopeptides from the tropical marine cyanobacterium Symploca sp. J. Nat. Prod. 2014, 77, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Al-Awadhi, F.H.; Ratnayake, R.; Paul, V.J.; Luesch, H. Tasiamide F, a potent inhibitor of cathepsins D and E from a marine cyanobacterium. Bioorg. Med. Chem. 2016, 24, 3276–3282. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.H.; Wray, V.; Nimtz, M.; Fossen, T.; Preisitsch, M.; Schröder, G.; Wende, K.; Heiden, S.E.; Mundt, S. Balticidins A–D, antifungal hassallidin-like lipopeptides from the Baltic Sea cyanobacterium Anabaena cylindrica Bio33. J. Nat. Prod. 2014, 77, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Vestola, J.; Shishido, T.K.; Jokela, J.; Fewer, D.P.; Aitio, O.; Permi, P.; Wahlsten, M.; Wang, H.; Rouhiainen, L.; Sivonen, K. Hassallidins, antifungal glycolipopeptides, are widespread among cyanobacteria and are the end-product of a nonribosomal pathway. Proc. Natl. Acad. Sci. USA 2014, 111, E1909–E1917. [Google Scholar] [CrossRef] [PubMed]
- Neuhof, T.; Schmieder, P.; Preussel, K.; Dieckmann, R.; Pham, H.; Bartl, F.; von Döhren, H. Hassallidin A, a glycosylated lipopeptide with antifungal activity from the cyanobacterium Hassallia sp. J. Nat. Prod. 2005, 68, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Ohno, O.; Sumimoto, S.; Suda, S.; Suenaga, K. Kurahyne, an acetylene-containing lipopeptide from a marine cyanobacterial assemblage of Lyngbya sp. RSC Adv. 2014, 4, 12840–12843. [Google Scholar] [CrossRef]
- Okamoto, S.; Iwasaki, A.; Ohno, O.; Suenaga, K. Isolation and Structure of Kurahyne B and Total Synthesis of the Kurahynes. J. Nat. Prod. 2015, 78, 2719–2725. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Ohno, O.; Sumimoto, S.; Ogawa, H.; Nguyen, K.A.; Suenaga, K. Jahanyne, an apoptosis-inducing lipopeptide from the marine cyanobacterium Lyngbya sp. Org. Lett. 2015, 17, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Sumimoto, S.; Iwasaki, A.; Ohno, O.; Sueyoshi, K.; Teruya, T.; Suenaga, K. Kanamienamide, an Enamide with an Enol Ether from the Marine Cyanobacterium Moorea bouillonii. Org. Lett. 2016, 18, 4884–4887. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Tadenuma, T.; Sumimoto, S.; Ohshiro, T.; Ozaki, K.; Kobayashi, K.; Teruya, T.; Tomoda, H.; Suenaga, K. Biseokeaniamides A, B, and C, Sterol O-Acyltransferase Inhibitors from an Okeania sp. Marine Cyanobacterium. J. Nat. Prod. 2017, 80, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Zhou, W.; Othman, R.; Uchida, H.; Watanabe, R.; Suzuki, T.; Sakamoto, B.; Nagai, H. A new malyngamide from the marine cyanobacterium Moorea producens. Nat. Prod. Res. 2017, 6419, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sabry, O.M.; Goeger, D.E.; Gerwick, W.H. Biologically active new metabolites from a Florida collection of Moorea producens. Nat. Prod. Res. 2017, 31, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Shaala, L.A.; Youssef, D.T.A.; McPhail, K.L.; Elbandy, M. Malyngamide 4, a new lipopeptide from the Red Sea marine cyanobacterium Moorea producens (formerly Lyngbya majuscula). Phytochem. Lett. 2013, 6, 183–188. [Google Scholar] [CrossRef]
- Quintana, J.; Bayona, L.M.; Castellanos, L.; Puyana, M.; Camargo, P.; Aristizábal, F.; Edwards, C.; Tabudravu, J.N.; Jaspars, M.; Ramos, F.A. Almiramide D, cytotoxic peptide from the marine cyanobacterium Oscillatoria nigroviridis. Bioorg. Med. Chem. 2014, 22, 6789–6795. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Iwasaki, A.; Sumimoto, S.; Iwatsuki, M.; Ishiyama, A.; Hokari, R.; Otoguro, K.; Omura, S.; Suenaga, K. Isolation and Total Synthesis of Hoshinolactam, an Antitrypanosomal Lactam from a Marine Cyanobacterium. Org. Lett. 2017, 19, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Bertin, M.J.; Wahome, P.G.; Zimba, P.V.; He, H.; Moeller, P.D.R. Trichophycin A, a cytotoxic linear polyketide isolated from a Trichodesmium thiebautii bloom. Mar. Drugs 2017, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Bertin, M.J.; Demirkiran, O.; Navarro, G.; Moss, N.A.; Lee, J.; Goldgof, G.M.; Vigil, E.; Winzeler, E.A.; Valeriote, F.A.; Gerwick, W.H. Kalkipyrone B, a marine cyanobacterial γ-pyrone possessing cytotoxic and anti-fungal activities. Phytochemistry 2016, 122, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Kinnel, R.B.; Esquenazi, E.; Leao, T.; Moss, N.; Mevers, E.; Pereira, A.R.; Monroe, E.A.; Korobeynikov, A.; Murray, T.F.; Sherman, D.; et al. A Maldiisotopic approach to discover natural products: Cryptomaldamide, a hybrid tripeptide from the marine cyanobacterium Moorea producens. J. Nat. Prod. 2017, 80, 1514–1521. [Google Scholar] [CrossRef] [PubMed]
- Balunas, M.J.; Grosso, M.F.; Villa, F.A.; Engene, N.; McPhail, K.L.; Tidgewell, K.; Pineda, L.M.; Gerwick, L.; Spadafora, C.; Kyle, D.E.; et al. Coibacins A–D, antileishmanial marine cyanobacterial polyketides with intriguing biosynthetic origins. Org. Lett. 2012, 14, 3878–3881. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.L.; Linington, R.G.; Balunas, M.J.; Centeno, A.; Boudreau, P.; Zhang, C.; Engene, N.; Spadafora, C.; Mutka, T.S.; Kyle, D.E.; et al. Bastimolide A, a Potent Antimalarial Polyhydroxy Macrolide from the Marine Cyanobacterium Okeania hirsuta. J. Org. Chem. 2015, 80, 7849–7855. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Reyes, L.A.; Sneed, J.; Paul, V.J.; Luesch, H. Amantelides A and B, Polyhydroxylated Macrolides with Differential Broad-Spectrum Cytotoxicity from a Guamanian Marine Cyanobacterium. J. Nat. Prod. 2015, 78, 1957–1962. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Williams, H.; Cagle, D.; Karanovich, K.; Horgen, F.D.; Smith, R.; Watanabe, C.M.H. Macrolactone nuiapolide, isolated from a Hawaiian marine cyanobacterium, exhibits anti-chemotactic activity. Mar. Drugs 2015, 13, 6274–6290. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Iwasaki, A.; Sumimoto, S.; Kanamori, Y.; Ohno, O.; Iwatsuki, M.; Ishiyama, A.; Hokari, R.; Otoguro, K.; Omura, S.; et al. Janadolide, a cyclic polyketide-peptide hybrid possessing a tert-butyl group from an Okeania sp. marine cyanobacterium. J. Nat. Prod. 2016, 79, 1862–1866. [Google Scholar] [CrossRef] [PubMed]
- Navarro, G.; Cummings, S.; Lee, J.; Moss, N.; Glukhov, E.; Valeriote, F.A.; Gerwick, L.; Gerwick, W.H. Isolation of Polycavernoside D from a Marine Cyanobacterium. Environ. Sci. Technol. Lett. 2015, 2, 166–170. [Google Scholar] [CrossRef]
- Gupta, D.K.; Kaur, P.; Leong, S.T.; Tan, L.T.; Prinsep, M.R.; Chu, J.J.H. Anti-Chikungunya viral activities of aplysiatoxin-related compounds from the marine cyanobacterium Trichodesmium erythraeum. Mar. Drugs 2014, 12, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Naman, C.B.; Engene, N.; Gerwick, W.H. Laucysteinamide a, a hybrid PKS/NRPS metabolite from a Saipan Cyanobacterium, cf. Caldora penicillata. Mar. Drugs 2017, 15, 121. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.; Engene, N.; Gunasekera, S.P.; Sneed, J.M.; Paul, V.J. Carriebowlinol, an antimicrobial tetrahydroquinolinol from an assemblage of marine cyanobacteria containing a novel taxon. J. Nat. Prod. 2015, 78, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Krunic, A.; Lantvit, D.; Shen, Q.; Kroll, D.J.; Swanson, S.M.; Orjala, J. Nitrile-containing fischerindoles from the cultured cyanobacterium Fischerella sp. Tetrahedron 2012, 68, 3205–3209. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lantvit, D.; Hwang, C.H.; Kroll, D.J.; Swanson, S.M.; Franzblau, S.G.; Orjala, J. Indole alkaloids from two cultured cyanobacteria, Westiellopsis sp. and Fischerella muscicola. Bioorg. Med. Chem. 2012, 20, 5290–5295. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.; Krunic, A.; Santarsiero, B.D.; Franzblau, S.G.; Orjala, J. Hapalindole-related alkaloids from the cultured cyanobacterium Fischerella ambigua. Phytochemistry 2010, 71, 2116–2123. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, M.; Pereira, A.R.; Debonsi, H.M.; Ligresti, A.; Di Marzo, V.; Gerwick, W.H. Cannabinomimetic lipid from a marine cyanobacterium. J. Nat. Prod. 2011, 74, 2313–2317. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Matthews, J.H.; Paul, V.J.; Luesch, H. Pitiamides A and B, Multifunctional Fatty Acid Amides from Marine Cyanobacteria. Planta Med. 2016, 82, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Kleigrewe, K.; Almaliti, J.; Tian, I.Y.; Kinnel, R.B.; Korobeynikov, A.; Monroe, E.A.; Duggan, B.M.; Di Marzo, V.; Sherman, D.H.; Dorrestein, P.C.; et al. Combining Mass Spectrometric Metabolic Profiling with Genomic Analysis: A Powerful Approach for Discovering Natural Products from Cyanobacteria. J. Nat. Prod. 2015, 78, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Leao, P.N.; Nakamura, H.; Costa, M.; Pereira, A.R.; Martins, R.; Vasconcelos, V.; Gerwick, W.H.; Balskus, E.P. Biosynthesis-Assisted Structural Elucidation of the Bartolosides, Chlorinated Aromatic Glycolipids from Cyanobacteria. Angew. Chem. Int. Ed. 2015, 54, 11063–11067. [Google Scholar] [CrossRef] [PubMed]
- Afonso, T.B.; Costa, M.S.; Rezende De Castro, R.; Freitas, S.; Silva, A.; Schneider, M.P.C.; Martins, R.; Leão, P.N. Bartolosides E–K from a marine coccoid cyanobacterium. J. Nat. Prod. 2016, 79, 2504–2513. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Fatma, T. Production of Indole-3-Acetic Acid by Cyanobacterial Strains. Nat. Prod. J. 2017, 7, 112–120. [Google Scholar] [CrossRef]
- Tarman, K.; Lindequist, U.; Mundt, S. Metabolites of marine microorganisms and their pharmacological activities. Mar. Microbiol. Bioact. Compd. Biotechnol. Appl. 2013, 393–416. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, T.; Ma, Z.; Li, Y. Design, synthesis and biological evaluation of tasiamide analogues as tumor inhibitors. Mar. Drugs 2014, 12, 2308–2325. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, W.; Xu, Y.; Ren, S.; Zhang, W.; Li, Y. Design, synthesis and biological evaluation of tasiamide B derivatives as BACE1 inhibitors. Bioorg. Med. Chem. 2015, 23, 1963–1974. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Singh, S. Characterization of NaCl-tolerant Mutant Strain of the Cyanobacterium Spirulina platensis Overproducing Phycocyanin. Nat. Prod. J. 2017, 7, 153–164. [Google Scholar] [CrossRef]
- Mandal, S.; Rath, J. Extremophilic Cyanobacteria for Novel Drug Development; Springer: Berlin, Germany, 2014; ISBN 3319120093. [Google Scholar]
- Sato, M.; Sagawa, M.; Nakazato, T.; Ikeda, Y.; Kizaki, M. A natural peptide, dolastatin 15, induces G2/M cell cycle arrest and apoptosis of human multiple myeloma cells. Int. J. Oncol. 2007, 30, 1453–1459. [Google Scholar] [CrossRef] [PubMed]
- Stratmann, K.; Moore, R.E.; Bonjouklian, R.; Deeter, J.B.; Patterson, G.M.L.; Shaffer, S.; Smith, C.D.; Smitka, T.A. Welwitindolinones, unusual alkaloids from the blue-green algae Hapalosiphon welwitschii and Westiella intricata. Relationship to fischerindoles and hapalinodoles. J. Am. Chem. Soc. 1994, 116, 9935–9942. [Google Scholar] [CrossRef]
- Nagle, D.G.; Park, P.U.; Paul, V.J. Pitiamide A, a new chlorinated lipid from a mixed marine cyanobacterial assemblage. Tetrahedron Lett. 1997, 38, 6969–6972. [Google Scholar] [CrossRef]
- Ribe, S.; Kondru, R.K.; Beratan, D.N.; Wipf, P. Optical rotation computation, total synthesis, and stereochemistry assignment of the marine natural product pitiamide A. J. Am. Chem. Soc. 2000, 122, 4608–4617. [Google Scholar] [CrossRef]
- Blake, D.R.; Robson, P.; Ho, M.; Jubb, R.W.; McCabe, C.S. Preliminary assessment of the efficacy, tolerability and safety of a cannabis-based medicine (Sativex) in the treatment of pain caused by rheumatoid arthritis. Rheumatology 2005, 45, 50–52. [Google Scholar] [CrossRef] [PubMed]






| Compounds | Species | Bioactivity | References |
|---|---|---|---|
| Urumamide (1) | Okeania sp. | Anticancer | [32] |
| Medusamide A (2) | Cyanobacterial sp. | No cytotoxicity | [33] |
| Odoamide (3) | Okeania sp. | Brine shrimp toxicity | [34] |
| Bouillonamide (4) | Moorea bouillonii | Antitumor | [35] |
| Companeramides A–B (5–6) | Cyanobacterial assemblage | Antiplasmodium | [36] |
| Dudawalamides A–D (7–10) | Moorea producens | Antiplasmodium and Antiparasitic | [37] |
| Kohamamides A–C (11–13) | Okeania sp. | Weak against Cancer | [38] |
| Kohamamide B (12) | Okeania sp. | Potent cytotoxicity | [38] |
| Viequeamide A–B (14–15) | Rivularia sp. | Potent anticancer activity | [39] |
| Pitiprolamide (16) | Lyngbya majuscula | Weak cytotoxic activity | [40] |
| Pitipeptolide C–E (17–19) | Lyngbya majuscula | Weak cytotoxic activity | [41] |
| Pitipeptolide (F) (20) | Lyngbya majuscula | Potent microbial activity | [41] |
| Apratoxin H (21) | Moorea producens | Anticancer activity | [42] |
| Apratoxin A sulfoxide (22) | Moorea producens | No cytoxicity | [42] |
| Samoamide A (23) | Symploca sp. | Anticancer activity | [43] |
| Wewakzole B (24) | Moorea producens | Anticancer activity | [44] |
| Odobromoamide (25) | Okeania sp. | Antitumor activity | [46] |
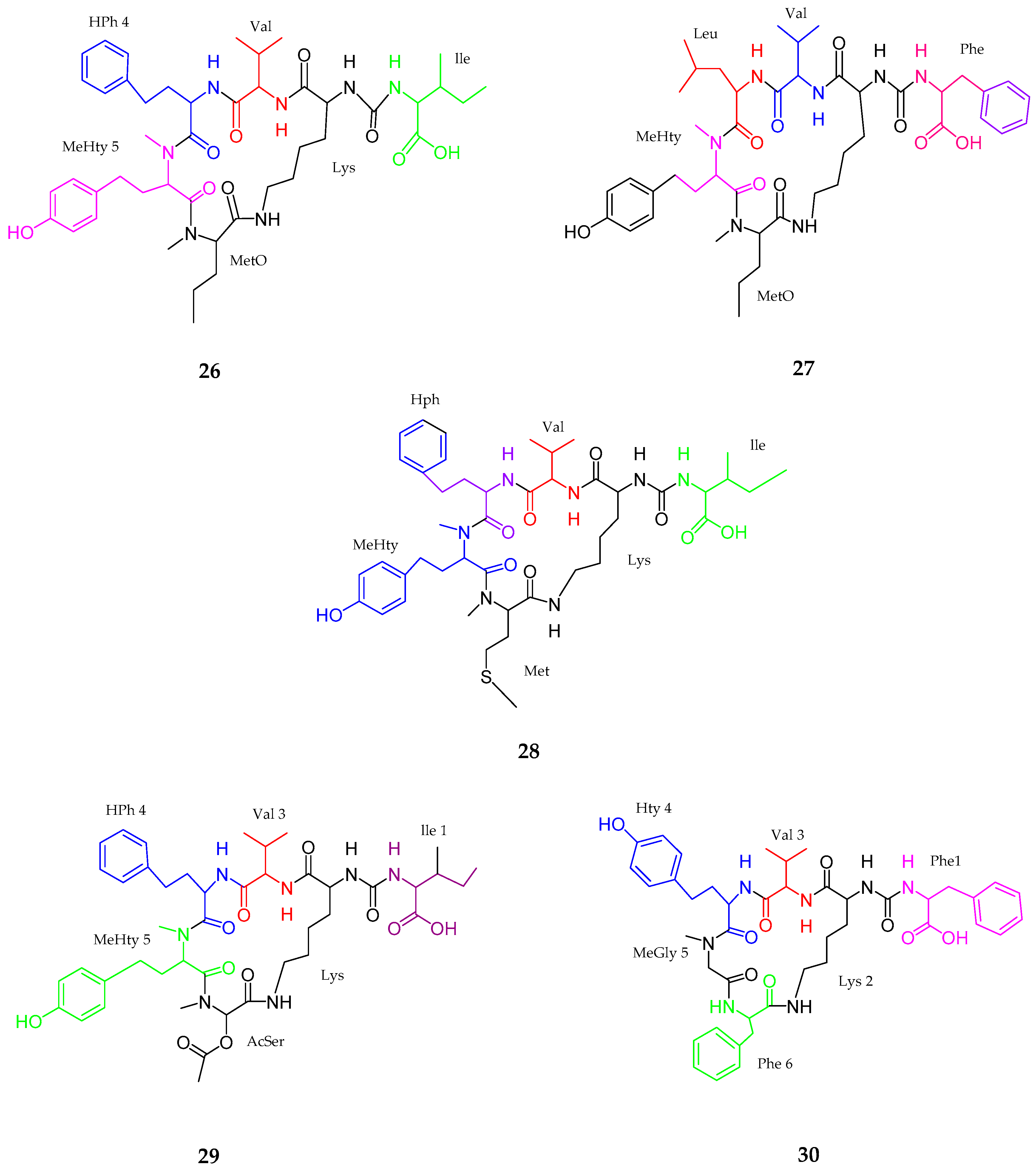
| Anabaenopeptins NP 883, 869, 867, 865, 813 (26–30) | Cyanobacterial sp. | No cytotoxicity | [47] |
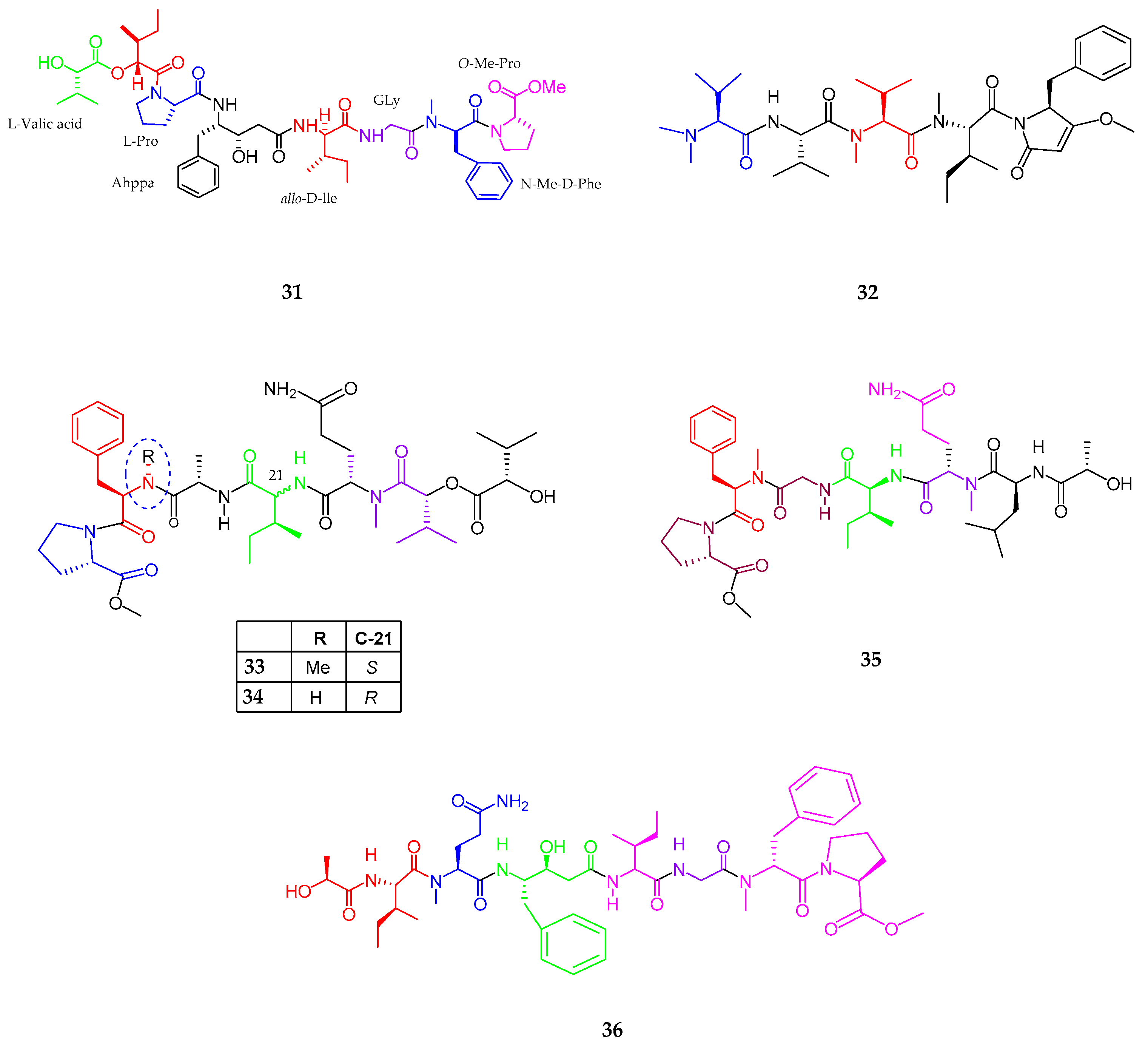
| Maedamide (31) | Lyngbya sp. | Strong anti-chymotrypsin | [48] |
| Caldoramide (32) | Caldora penicillata | Antitumor | [49] |
| Tasiamides C–E (33–35) | Symploca sp. | No cytotoxicity | [50] |
| Tesiamide F (36) | Lyngbya sp. | Anti-proteolytic activity | [51] |
| Balticidin A–D (37–40) | Anabaena cylindrica | Antifungal | [52] |
| Hassallidins C–D (41–42) | Anabaena sp. | Anti- fungal | [53] |
| Kurahyne (43) | Lyngbya sp. | Anti-cancer | [55] |
| Kurahyne B (44) | Lyngbya sp. | Anti-cancer | [56] |
| Jahanyne (45) | Lyngbya sp. | Anti-tumor activity | [57] |
| Kanamienamide (46) | Moorea bouillonii | Antitumor | [58] |
| Biseokeaniamides A–C (47–49) | Okeania sp. | inhibited sterol O-acyltransferase SOAT1 and SOAT2 | [59] |
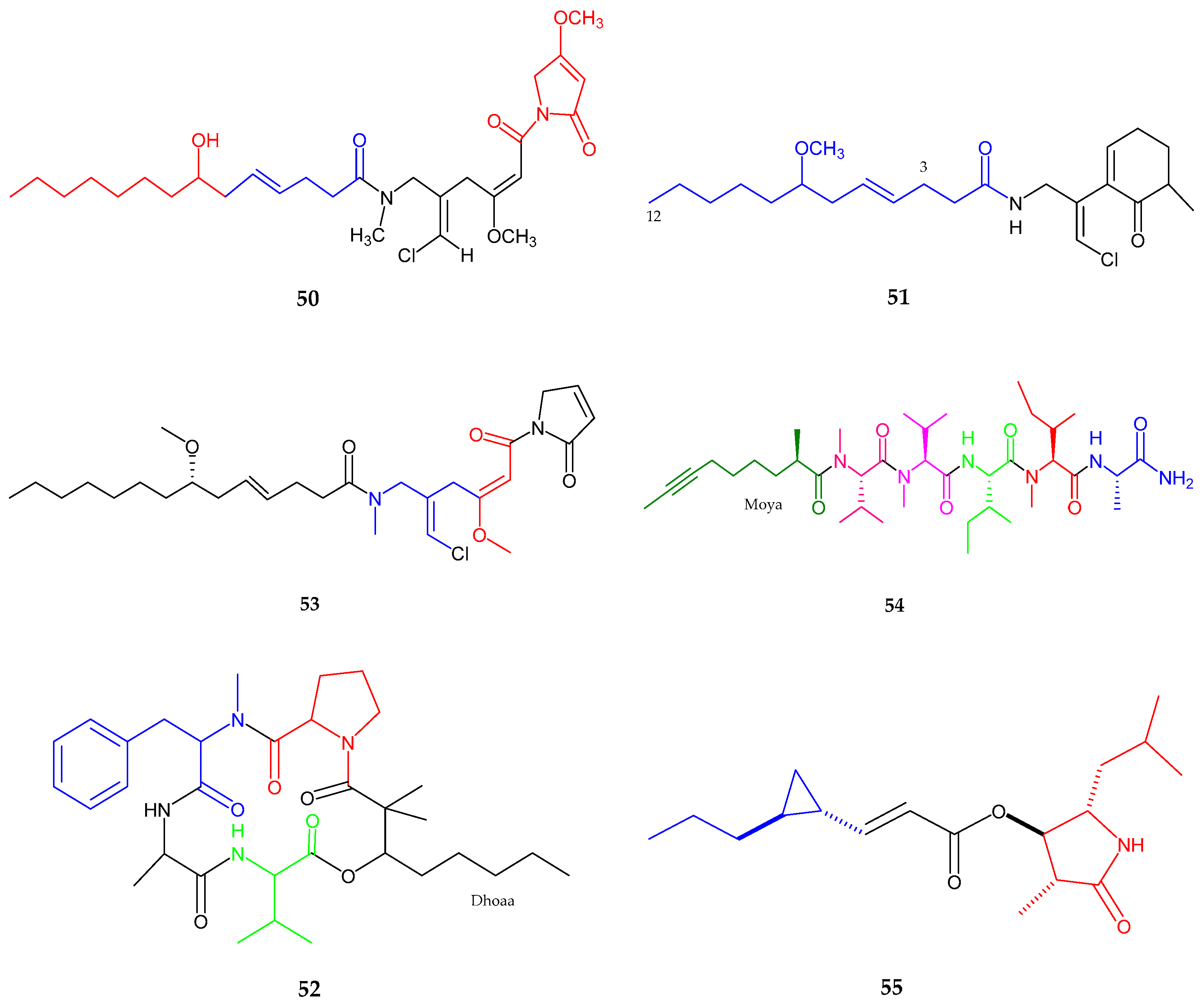
| Maylngamide (50) | Moorea producens | Cytotoxic | [60] |
| Malyngamide Y, (+)-floridamide (51–52) | Moorea producens | Anticancer | [61] |
| Malyngamide 4 (53) | Moorea producens | Anticancer | [62] |
| Almiramide D (54) | Oscillatoria nigroviridis | Potent brine shirimp toxicity | [63] |
| Hoshinolactam (55) | Cyanobacterial sp. | Anti-trypanosomal activity | [64] |
| Compound | Species | Bioactivities | References |
|---|---|---|---|
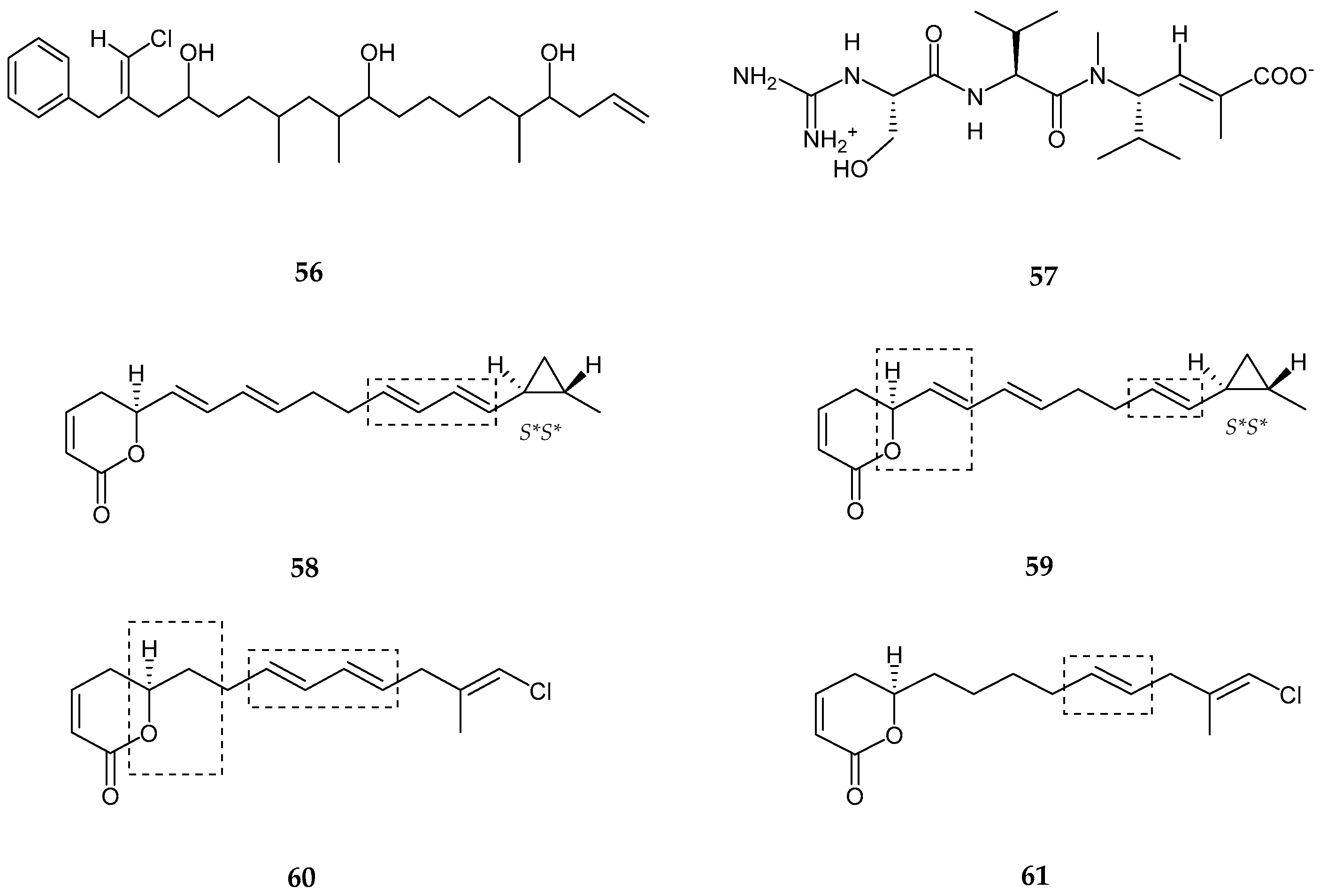
| Trichophycin A (56) | Trichodesmium thiebautii | Anti-cancer | [65] |
| Cryptomaldamide (57) | Moorea producens | No Cytotoxicity | [67] |
| Coibacins A–D (58–61) | Oscillatoria sp. | Anti-parasite, cancer and inflammatory | [68] |
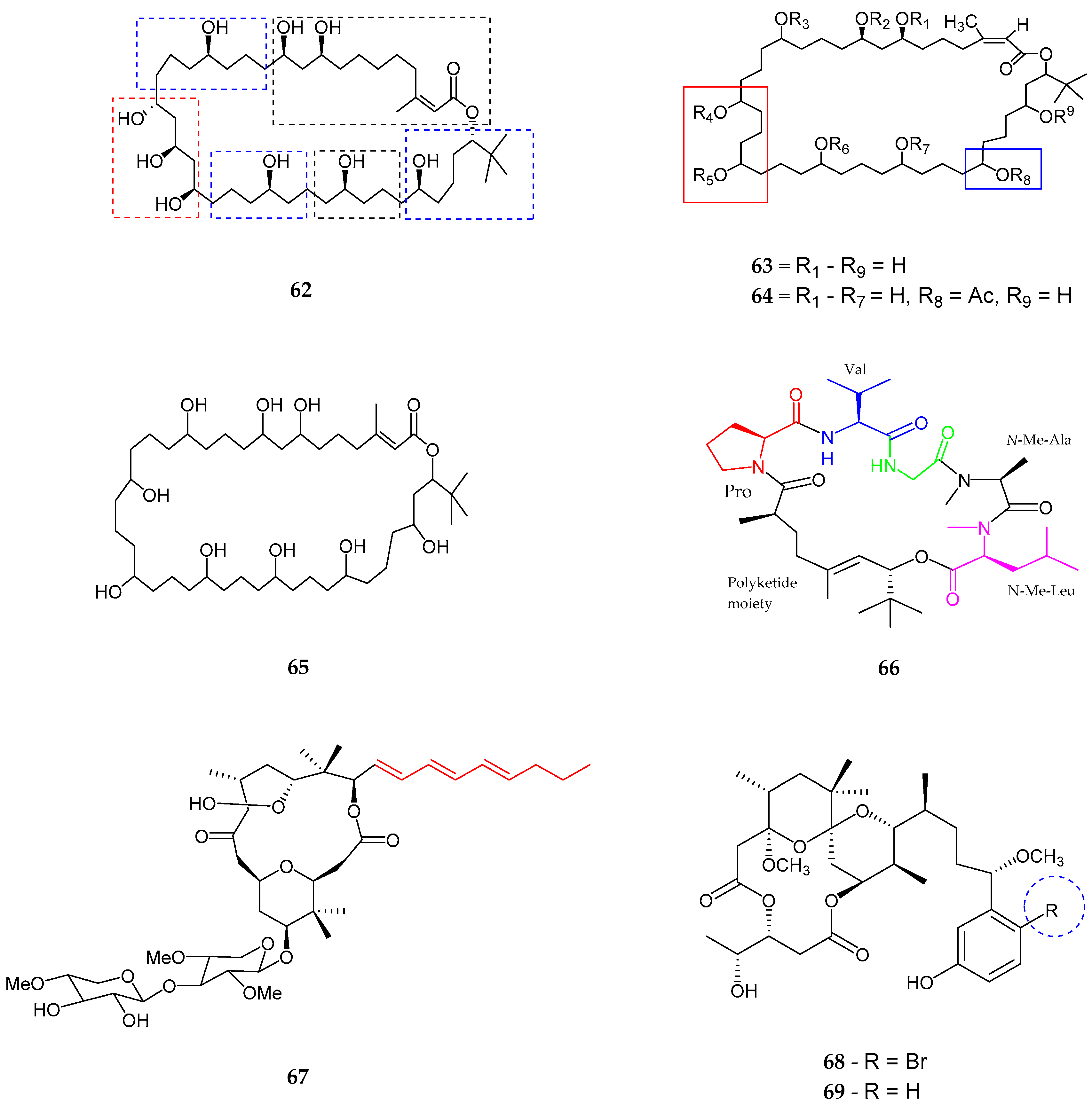
| Bastimolide A (62) | Okeania hirsuta | Antimalarial activity | [69] |
| Amentelides A–B (63–64) | Gray cyanobacterium | Anti-Cancer | [70] |
| Nuiapolide (65) | Okeania plumata | Anti-cancer | [71] |
| Janadolide (66) | Okeania sp. | Anti-trypanasomal against Trypanosoma brucei brucei | [72] |
| Polycavernoside D (67) | Okeania sp. | Anti-cancer | [73] |
| 3-methoxyaplysiatoxin and 3-methoxydebromoaplysiatoxin (68–69) | Trichodesmium erythraeum | Anti-viral | [74] |
| Compounds | Species | Bioactivities | References |
|---|---|---|---|
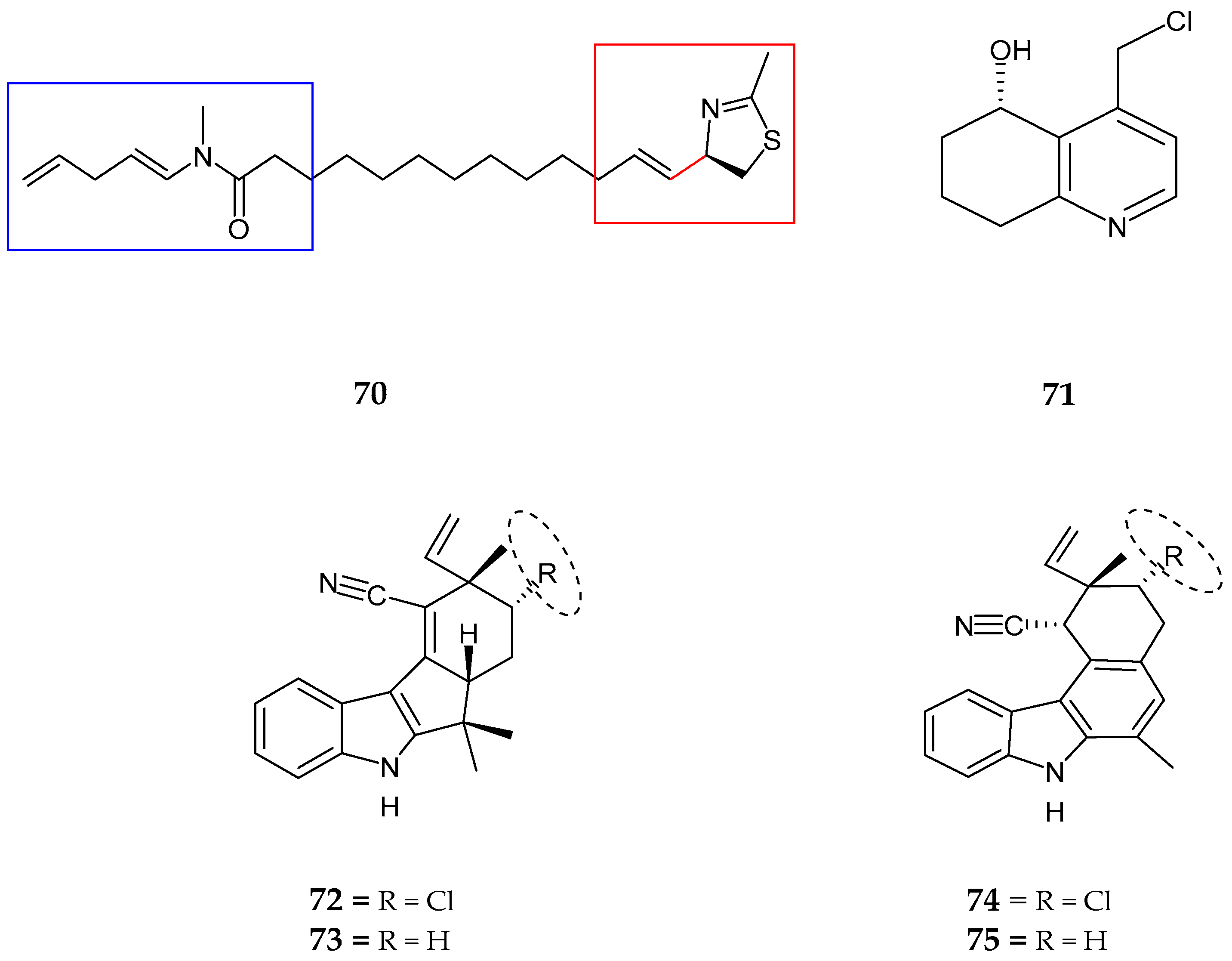
| Laucysteinamide A (70) | Caldora Penicillata | Anticancer | [75] |
| Carriebowlinol (71) | Cyanobacterial mat | Anti-fungal | [76] |
| 12-epi-fischerindole I (72) | Fischerella sp. | No cytotoxicity | [77] |
| Deschloro 12-epi-fischerindole I (73) | Fischerella sp. | Anti-cancer | [77] |
| 12-epi-fischerindole W (74) | Fischerella sp. | No cytotoxicity | [77] |
| Deschloro 12-epi-fischerindole W (75) | Fischerella sp. | No cytotoxicity | [77] |
| hapalindole X (76) | Westiellopsis sp. | Anti-cancer and anti-microbial | [78] |
| deschloro hapalindole I (77) | Fischerella muscicola | Anti-cancer and anti-microbial | [78] |
| 13-hydroxydechlorofontonamide (78) | Westiellopsis sp. and Fischerella muscicola | Anti-cancer and anti-microbial | [78] |
| fischambiguines A–B (79–80), Ambiguine P (81) and Ambiguine Q (82) | Fischerella ambigua. | Anti-tuberculosis | [79] |
| Compound | Species | Bioactivity | References |
|---|---|---|---|
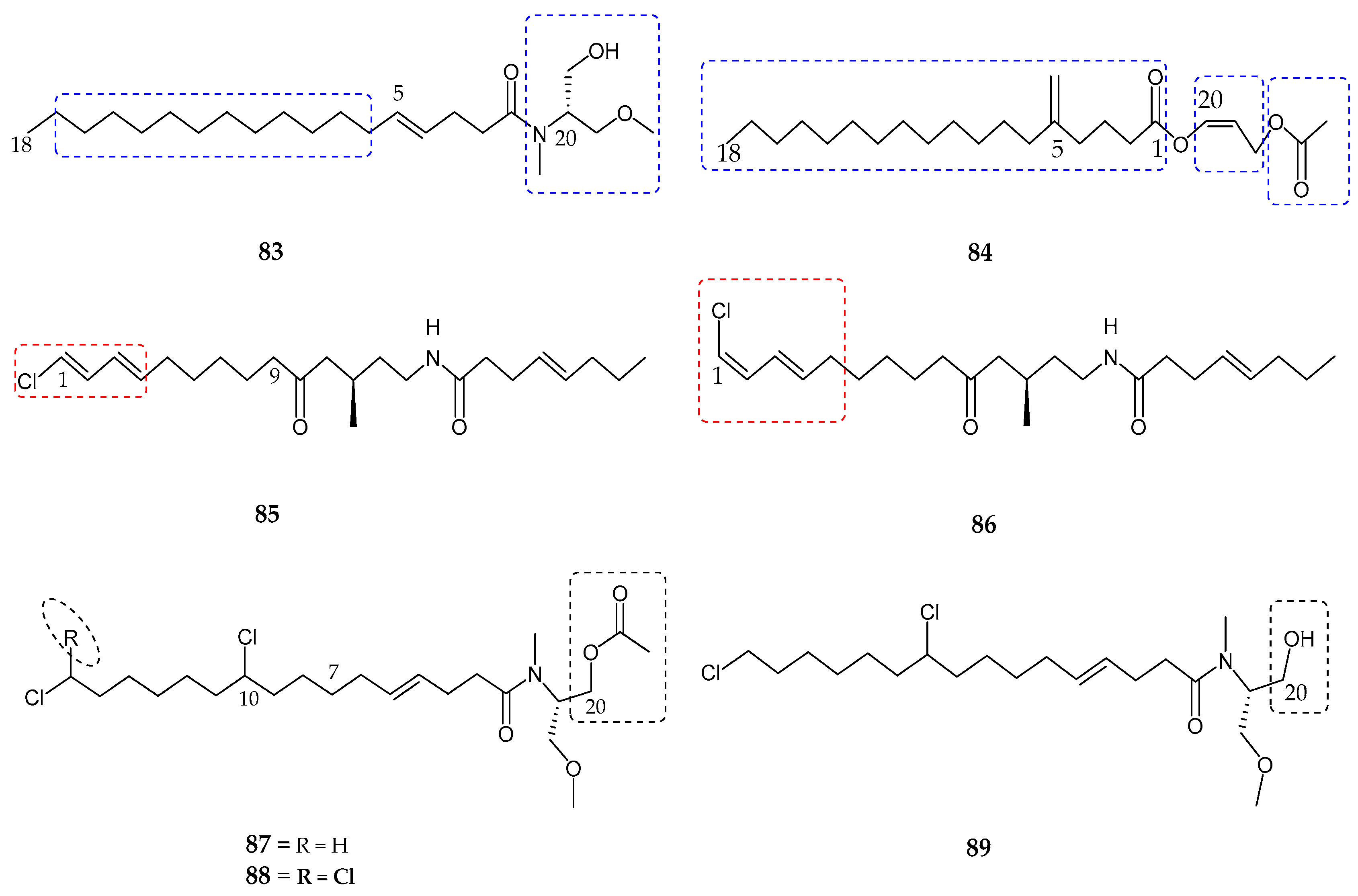
| Serinolamide A (83) | Lyngbia majuscula | Moderate affinity for CB1 receptor | [80] |
| Propenediester (84) | Oscillatoria sp. | No affinity | [80] |
| 1E-pitiamide B (85) | Cyanobacterial sp. | Anti-cancer | [81] |
| 1Z-pitiamide B (86) | Cyanobacterial sp. | Anti-cancer | [81] |
| Columbamides A–C (87–89) | Cyanobacterial sp. | Potent affinity for CB1 and CB2 receptors | [82] |
| Bartolosides A–D (90–93) | Nodosilinea sp. and Synechocystis salina sp. | Not reported | [83] |
| Bartolosides E–K (94–100) | Synechocystis Salina | No cytotoxicity | [84] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, S.A.A.; Akhter, N.; Auckloo, B.N.; Khan, I.; Lu, Y.; Wang, K.; Wu, B.; Guo, Y.-W. Structural Diversity, Biological Properties and Applications of Natural Products from Cyanobacteria. A Review. Mar. Drugs 2017, 15, 354. https://doi.org/10.3390/md15110354
Shah SAA, Akhter N, Auckloo BN, Khan I, Lu Y, Wang K, Wu B, Guo Y-W. Structural Diversity, Biological Properties and Applications of Natural Products from Cyanobacteria. A Review. Marine Drugs. 2017; 15(11):354. https://doi.org/10.3390/md15110354
Chicago/Turabian StyleShah, Sayed Asmat Ali, Najeeb Akhter, Bibi Nazia Auckloo, Ishrat Khan, Yanbin Lu, Kuiwu Wang, Bin Wu, and Yue-Wei Guo. 2017. "Structural Diversity, Biological Properties and Applications of Natural Products from Cyanobacteria. A Review" Marine Drugs 15, no. 11: 354. https://doi.org/10.3390/md15110354






