New 9α-Hydroxy-5α,6α-epoxysterols from the Vietnamese Marine Sponge Ircinia echinata
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Materials
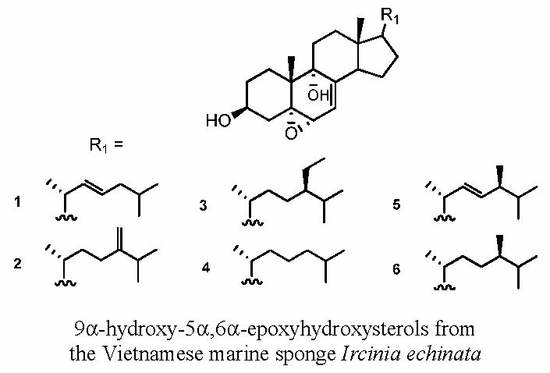
3.3. Isolation of 5α,6α-Epoxysterols from the Vietnamese Marine Sponge Ircinia echinata
3.3.1. 5α,6α-Epoxycholesta-7,22(E)-dien-3β,9α-diol (1)
3.3.2. 5α,6α-Epoxycholesta-7,24(28)-dien-3β,9α-diol (2)
3.3.3. (24R)-5α,6α-Epoxy-24-ethyl-cholesta-7-en-3β,9α-diol (3)
3.3.4. 5α,6α-Epoxycholesta-7-en-3β,9α-diol (4)
3.3.5. (24S)-5α,6α-Epoxyergosta-7,22-dien-3β,9α-diol (5)
3.3.6. (24R)-5α,6α-Epoxy-24-methyl-cholesta-7-en-3β,9α-diol (6)
3.4. Evaluation of Cytotoxic Activity
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kawakami, A.; Miyamoto, T.; Higuchi, R.; Uchiumi, T.; Kuwano, M.; Soet, R.W.M.V. Structure of a novel multidrug resistance modulator, irciniasulfonic acid, isolated from a marine sponge Ircinia sp. Tetrahedron Lett. 2001, 42, 3335–3337. [Google Scholar] [CrossRef]
- Sica, D.; Piccialli, V.; Pronzato, R. Sterols from the sponges Ircinia pipetta & Dysidea avara. Identification of cholestatrienol. Comp. Biochem. Physiol. 1987, 88, 293–296. [Google Scholar]
- Venkateswarlu, Y.; Reddy, M.V.R.; Rao, M.N. A new epoxy sterol from the sponge Ircinia fasciculata. J. Nat. Prod. 1996, 59, 876–877. [Google Scholar] [CrossRef]
- Hahn, D.; Chin, J.; Kim, H.; Yang, I.; Won, D.H.; Ekins, M.; Choi, H.; Nam, S.J.; Kang, H. Sesquiterpenoids with PPARδ agonistic effect from a Korean marine sponge Ircinia sp. Tetrahedron Lett. 2007, 55, 4716–4719. [Google Scholar] [CrossRef]
- Buchanan, M.S.; Edser, A.; King, G.; Whitmore, J.; Quin, R.J. Cheilanthane sesterterpenes, protein kinase inhibitors, from a marine sponge of the genus Ircinia. J. Nat. Prod. 2001, 64, 300–303. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.-Y.; Lu, M.-C.; Wang, L.-H.; Chen, J.-J.; Fang, L.-S.; Wu, Y.-C.; Sung, P.-J. New scalarane sesterterpenoids from the Formosan sponge Ircinia felix. Mar. Drugs 2015, 13, 4296–4309. [Google Scholar] [CrossRef] [PubMed]
- Issa, H.H.; Tanaka, J.; Higa, T. New cytotoxic furanosesterterpenes from an Okinawan marine sponge Ircinia sp. J. Nat. Prod. 2003, 66, 252–254. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Caroll, A.R.; Pass, D.M.; Archbold, J.K.; Avery, V.M.; Quin, R.J.J. Polydiscamides B−D from a marine sponge Ircinia sp. as potent human sensory neuron-specific G protein coupled receptor agonists. J. Nat. Prod. 2008, 71, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.A.; Gustafson, K.R.; Boyd, M.R. New chondropsin macrolide lactams from marine sponges in the genus Ircinia. Tetrahedron Lett. 2001, 42, 1623–1626. [Google Scholar] [CrossRef]
- Chevallier, C.; Bugni, T.S.; Feng, X.; Harper, M.K.; Orendt, A.M.; Ireland, C.M. Tedanolide C: A potent new 18-membered-ring cytotoxic macrolide isolated from the Papua New Guinea marine sponge Ircinia sp. J. Org. Chem. 2006, 71, 2510–2513. [Google Scholar] [CrossRef] [PubMed]
- Marquez, F.D.M.; Martinez, M.A. Antileishmanial epidioxysterols from the Colombian marine sponge Ircinia campana are oxidation products from naturally occurring Δ5,7 sterols. Vitae 2007, 14, 61–66. [Google Scholar]
- Xu, S.; Liao, X.; Du, B.; Zhou, X.; Huang, Q.; Wu, C. A series of new 5,6-epoxysterols from a Chinese sponge Ircinia aruensis. Steroids 2008, 73, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Shinonaga, H.; Shigemori, H.; Umeyama, A.; Shoji, N.; Arihara, S. Xestobergsterol C, a new pentacyclic steroid from the Okinawan marine sponge Ircinia sp. and absolute stereochemistry of Xestobergsterol A. J. Nat. Prod. 1995, 58, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.; Choi, H.; Won, D.H.; Nam, S.-J.; Kang, H. An antibacterial 9,11-secosterol from a marine sponge Ircinia sp. Bull. Korean Chem. Soc. 2014, 35, 3360–3362. [Google Scholar] [CrossRef]
- Yang, I.; Choi, H.; Nam, S.-J.; Kang, H. A new 9,11-secosterol with a 1,4-quinone from a Korean marine sponge Ircinia sp. Arch. Pharm. Res. 2015, 38, 1970–1974. [Google Scholar] [CrossRef] [PubMed]
- Do, T.T.; Duong, T.D.; Do, C.T.; Pham, H.Y.; Nguyen, X.N.; Dan, T.T.H.; Bui, H.T.; Hoang, L.T.A.; Nguyen, T.C.; Chau, V.M.; et al. Sterols from the Vietnamese sponge Ircinia echinata. Vietnam J. Chem. 2016, 54, 72–76. [Google Scholar]
- Phan, V.K.; Duong, T.D.; Do, T.T.; Tran, H.Q.; Nguyen, T.T.N.; Tran, M.H.; Hoang, L.T.A.; Pham, H.Y.; Do, T.T.; Nguyen, X.N.; et al. Constituents from Ircinia echinata and their antiproliferative effect on six human cancer cell strains. Lett. Org. Chem. 2017, 14, 248–253. [Google Scholar]
- Vanderah, D.J.; Djerassi, C. Marine natural products. Synthesis of four naturaly occurring 20β-H Cholanic acid derivatives. J. Org. Chem. 1978, 43, 1442–1448. [Google Scholar] [CrossRef]
- Aiello, A.; Fattorusso, E.; Menna, M. Four new bioactive polyhydroxylated sterols from the black coral Antipathes subpinnata. J. Nat. Prod. 1992, 55, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, T.A.; Lee, Y.M.; Hong, J.; Lee, C.-O.; Im, K.S.; Jung, J.H. 5,6:8,9-Diepoxy and other cytotoxic sterol from the marine sponge Homaxinella sp. J. Nat. Prod. 2006, 69, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.L.C.; McInnes, A.G.; Shimizu, S.; Smith, D.G.; Walter, J.A. Identification of C-24 alkyl epimers of marine sterol by 13C nuclear magnetic resonance spectroscopy. Can. J. Chem. 1978, 56, 1898–1903. [Google Scholar]
- Ioannou, E.; Abdel-Razik, A.F.; Zervou, M.; Christofidis, D.; Alexi, X.; Vagias, C.; Alexis, M.N.; Roussis, V. 5α,8α-epidioxysterols from the gorgonian Eunicella cavolini and the ascidian Trididemnum inarmatum: Isolation and evaluation of their antiproliferative activity. Steroids 2009, 74, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Korade, Z.; Porter, N.A. Oxysterols from free radical chain oxidation of 7-dehydrocholesterol: Product and mechanistic studies. J. Am. Chem. Soc. 2010, 132, 2222–2232. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]




| No | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 2.20 dt (4.3, 13.5) | 2.20 dt (4.3, 13.5) | 2.20 dt (4.3, 13.8) | 2.20 dt (4.2, 13.8) | 2.20 dt (4.2, 13.8) | 2.20 dt (4.2, 13.8) |
| 1.35 m | 1.35 m | 1.36 m | 1.35 m | 1.36 m | 1.36 m | |
| 2 | 1.86 m | 1.86 m | 1.88 m | 1.85 m | 1.86 m | 1.86 m |
| 1.51 m | 1.50 m | 1.52 m | 1.52 m | 1.51 m | 1.51 m | |
| 3 | 4.00 m | 4.00 m | 4.00 m | 4.00 m | 4.0 m | 4.0 m |
| 4 | 1.66 m | 1.66 m | 1.66 m | 1.67 m | 1.66 m | 1.67 m |
| 2.11 dd (11.4, 13.8) | 2.12 dd (11.2, 13.5) | 2.12 dd (11.2, 13.8) | 2.12 dd (11.4, 13.2) | 2.12 dd (11.4, 13.2) | 2.12 dd (11.4, 13.8) | |
| 6 | 3.65 m | 3.64 m | 3.66 m | 3.66 m | 3.65 m | 3.65 m |
| 7 | 5.33 m | 5.33 m | 5.34 dd (1.8, 4.8) | 5.34 dd (2.4, 4.8) | 5.34 dd (2.4, 4.8) | 5.33 dd (1.8, 4.8) |
| 11 | 1.60 m | 1.97 m | 1.97 m | 1.97 m | 1.60 m | 1.60 m |
| 1.91 m | 1.59 m | 1.60 m | 1.60 m | 1.90 m | 1.97 m | |
| 12 | 1.61 m | 1.60 m | 1.59 m | 1.59 m | 1.61 m | 1.60 m |
| 1.83 m | 1.86 m | 1.86 m | 1.86 m | 1.83 m | 1.86 m | |
| 14 | 2.49 m | 2.50 m | 2.49 m | 2.50 m | 2.50 m | 2.49 m |
| 15 | 1.55 m | 1.61 m | 1.61 m | 1.61 m | 1.56 m | 1.61 m |
| 1.48 m | 1.51 m | 1.53 m | 1.53 m | 1.48 m | 1.52 m | |
| 16 | 1.34 m | 1.36 m | 1.35 m | 1.34 m | 1.34 m | 1.34 m |
| 1.80 m | 1.90 m | 1.91 m | 1.90 m | 1.80 m | 1.91 m | |
| 17 | 1.37 m | 1.36 m | 1.36 m | 1.34 m | 1.37 m | 1.34 m |
| 18 | 0.66 s | 0.65 s | 0.65 s | 0.65 s | 0.66 s | 0.65 s |
| 19 | 1.12 s | 1.12 s | 1.12 s | 1.12 s | 1.12 s | 1.12 s |
| 20 | 2.05 m | 1.45 m | 1.41 m | 1.41 m | 2.04 m | 1.41 m |
| 21 | 1.03 d (6.5) | 0.99 d (6.8) | 0.97 d (6.5) | 0.96 d (6.6) | 1.03 d (6.6) | 0.96 d (6.6) |
| 22 | 5.24 dd (8.0, 15.0) | 1.18 m, 1.60 m | 1.60 m, 1.07 m | 1.41 m, 1.14 m | 5.20 m | 1.11 m, 1.40 m |
| 23 | 5.32 m | 1.92 m, 2.13 m | 1.25 m | 1.40 m, 1.19 m | 5.21 m | 1.28 m, 1.53 m |
| 24 | 1.84 m | 0.96 m | 1.16 m | 1.85 m | 1.23 m | |
| 25 | 1.58 m | 2.24 m | 1.71 m | 1.53 m | 1.47 m | 1.55 m |
| 26 | 0.89 d (6.7) | 1.03 d (6.7) | 0.86 d (6.8) | 0.89 d (6.6) | 0.88 d (7.2) | 0.88 d (7.2) |
| 27 | 0.89 d (6.7) | 1.04 d (6.7) | 0.84 d (6.8) | 0.89 d (6.6) | 0.86 d (7.2) | 0.83 d (7.2) |
| 28 | 4.67 brs, 4.73 brs | 1.35 m, 1.18 m | 0.94 d (7.2) | 0.81 d (7.2) | ||
| 29 | 0.88 m |
| No | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 28.2 t | 28.2 t | 28.3 t | 28.2 t | 28.3 t | 28.3 t |
| 2 | 31.6 t | 31.6 t | 31.6 t | 31.6 t | 31.6 t | 31.6 t |
| 3 | 68.2 d | 68.2 d | 68.2 d | 68.2 d | 68.2 d | 68.2 d |
| 4 | 40.7 t | 40.7 t | 40.8 t | 40.7 t | 40.7 t | 40.7 t |
| 5 | 78.9 s | 78.9 s | 78.9 s | 78.9 s | 78.9 s | 78.9 s |
| 6 | 73.7 d | 73.7 d | 73.7 d | 73.7 d | 73.7 d | 73.7 d |
| 7 | 121.0 d | 121.1 d | 121.0 d | 121.0 d | 121.0 d | 121.0 d |
| 8 | 143.8 s | 143.8 s | 143.8 s | 143.8 s | 143.8 s | 143.8 s |
| 9 | 76.1 s | 76.0 s | 76.1 s | 76.0 s | 76.1 s | 76.1 s |
| 10 | 41.4 s | 41.4 s | 41.4 s | 41.4 s | 41.4 s | 41.4 s |
| 11 | 29.1 t | 28.9 t | 29.0 t | 28.9 t | 29.2 t | 29.0 t |
| 12 | 36.5 t | 36.6 t | 36.6 t | 36.6 t | 36.5 t | 36.5 t |
| 13 | 44.8 s | 44.9 s | 44.9 s | 44.9 s | 44.8 s | 44.8 s |
| 14 | 51.9 d | 51.8 d | 51.8 d | 51.8 d | 51.9 d | 51.8 d |
| 15 | 24.0 t | 24.1 t | 24.1 t | 24.1 t | 24.0 t | 24.0 t |
| 16 | 29.3 t | 29.1 t | 29.2 t | 29.1 t | 29.6 t | 29.1 t |
| 17 | 57.3 d | 57.4 d | 57.4 d | 57.5 d | 57.4 d | 57.5 d |
| 18 | 12.2 q | 12.0 q | 12.2 q | 12.0 q | 12.2 q | 12.0 q |
| 19 | 22.2 q | 22.2 q | 22.2 q | 22.2 q | 22.2 q | 22.2 q |
| 20 | 41.8 d | 37.3 d | 37.9 d | 37.5 d | 41.9 d | 37.5 d |
| 21 | 21.5 q | 19.3 q | 19.4 q | 19.3 q | 21.5 q | 19.3 q |
| 22 | 139.1 d | 35.8 t | 35.0 t | 37.2 t | 137.2 d | 35.0 t |
| 23 | 127.8 d | 32.1 t | 27.2 t | 24.9 t | 133.4 d | 31.4 t |
| 24 | 43.1 t | 157.3 s | 47.4 d | 40.6 t | 44.6 d | 40.3 d |
| 25 | 29.8 d | 34.9 d | 30.2 d | 29.2 d | 34.5 d | 33.7 d |
| 26 | 22.7 q | 22.3 q | 20.1 q | 22.9 q | 20.7 q | 20.5 q |
| 27 | 22.7 q | 22.4 q | 19.4 q | 23.2 q | 20.1 q | 18.5 q |
| 28 | 106.1 t | 24.1 t | 18.6 q | 15.8 q | ||
| 29 | 12.7 q |
| Compound | Human Cancer Cell Lines | ||
|---|---|---|---|
| MCF-7 | HepG-2 | Lu-1 | |
| 3 | 15.88 ± 1.36 | >32 | >32 |
| 4 | 15.88 ± 0.09 | 15.95 ± 0.20 | 22.92 ± 0.09 |
| Ellipticine | 0.34 ± 0.01 | 0.38 ± 0.05 | 0.41 ± 0.04 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trinh, T.T.V.; Truong, B.N.; Longeon, A.; Doan, T.M.H.; Deville, A.; Chau, V.M.; Pham, V.C.; Bourguet-Kondracki, M.-L. New 9α-Hydroxy-5α,6α-epoxysterols from the Vietnamese Marine Sponge Ircinia echinata. Mar. Drugs 2018, 16, 424. https://doi.org/10.3390/md16110424
Trinh TTV, Truong BN, Longeon A, Doan TMH, Deville A, Chau VM, Pham VC, Bourguet-Kondracki M-L. New 9α-Hydroxy-5α,6α-epoxysterols from the Vietnamese Marine Sponge Ircinia echinata. Marine Drugs. 2018; 16(11):424. https://doi.org/10.3390/md16110424
Chicago/Turabian StyleTrinh, Thi Thanh Van, Bich Ngan Truong, Arlette Longeon, Thi Mai Huong Doan, Alexandre Deville, Van Minh Chau, Van Cuong Pham, and Marie-Lise Bourguet-Kondracki. 2018. "New 9α-Hydroxy-5α,6α-epoxysterols from the Vietnamese Marine Sponge Ircinia echinata" Marine Drugs 16, no. 11: 424. https://doi.org/10.3390/md16110424