Limonoids Containing a C1–O–C29 Moiety: Isolation, Structural Modification, and Antiviral Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Identification of Natural Limonoids 1–5
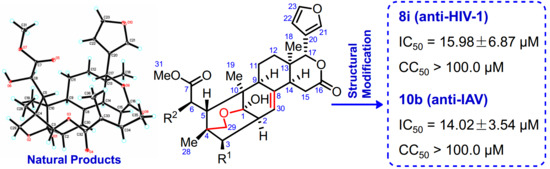
2.2. Structural Modifications of Granatumin L (6)
2.3. Antiviral Activity Bioassay
3. Materials and Methods
3.1. General
3.2. Plant Material
3.3. Extraction and Isolation
3.4. X-ray Crystal Data for Thaigranatin A (1)
3.5. Procedure for Structural Modification of Granatumin L (6)
3.5.1. General Procedure for the Preparation of 7
3.5.2. General Procedure for the Preparation of 8
3.5.3. General Procedure for the Preparation of 8a–8i
3.5.4. General Procedure for the Preparation of 9
3.5.5. General Procedure for the Preparation of 9a and 9b
3.5.6. General Procedure for the Preparation of 10
3.5.7. General Procedure for the Preparation of 10a–10c
3.6. Antiviral Bioassay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, J.; Xiao, Q.; Xu, J.; Li, M.Y.; Pan, J.Y.; Yang, M.H. Natural Products from True Mangrove Flora: Source, Chemistry and Bioactivities. Nat. Prod. Rep. 2008, 25, 955–981. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Li, X.W.; Guo, Y.W. Recent Progress on the Mangrove Plants: Chemistry and Bioactivity. Curr. Org. Chem. 2016, 20, 1923–1942. [Google Scholar] [CrossRef]
- Zhang, Q.; Satyanandamurty, T.; Shen, L.; Wu, J. Krishnolides A–D: New 2-Ketokhayanolides from the Krishna Mangrove, Xylocarpus moluccensis. Mar. Drugs 2017, 15, 333. [Google Scholar] [CrossRef] [PubMed]
- Li, W.S.; Jiang, Z.Z.; Shen, L.; Pedpradab, P.; Bruhn, T.; Wu, J.; Bringmann, G. Antiviral Limonoids Including Khayanolides from the Trang Mangrove Plant Xylocarpus moluccensis. J. Nat. Prod. 2015, 78, 1570–1578. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, S.; Xiao, Q.; Li, Q.X.; Huang, H.S.; Long, L.J.; Huang, L.M. Xlyogranatin L, a novel limonoid from Xylocarpus granatum. Tetrahedron. Lett. 2004, 45, 591–593. [Google Scholar] [CrossRef]
- Wu, J.; Ding, H.X.; Li, M.Y.; Zhang, S. Xylogranatin E, a New Phragmalin with a Rare Oxygen Bridge Between C1 and C29, from the Fruit of a Chinese Mangrove Xylocarpus granatum. Z. Naturforsch. 2007, 62, 569–572. [Google Scholar] [CrossRef]
- Yin, S.; Wang, X.N.; Fan, C.Q.; Lin, L.P.; Ding, J.; Yue, J.M. Limonoids from the Seeds of the Marine Mangrove Xylocarpus granatum. J. Nat. Prod. 2007, 70, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Xiao, Q.; Satyanandamurty, T.; Wu, J. Limonoids with an oxygen bridge between C(1) and C(29) from the seeds of a Krishna mangrove, Xylocarpus granatum. Chem. Biodivers. 2014, 11, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.G.; Wu, J.; Padmakumar, K.P.; Shen, L. Sundarbanxylogranins A-E, five new limonoids from the Sundarban Mangrove, Xylocarpus granatum. Fitoterapia 2017, 122, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.X.; Liao, Q.; Shen, L.; Wu, J. Krishnagranatins A-I: New limonoids from the mangrove, Xylocarpus granatum, and NF-κB inhibitory activity. Fitoterapia 2018, 131, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.H.; Pedpradab, P.; Wu, J. Thaixylogranins A–H: Eight new limonoids from the Thai mangrove, Xylocarpus granatum. Phytochem. Lett. 2017, 19, 126–131. [Google Scholar] [CrossRef]
- Li, J.; Li, M.Y.; Feng, G.; Xiao, Q.A.; Sinkkonen, J.; Satyanandamurty, T.; Wu, J. Limonoids from the seeds of Godavari mangrove, Xylocarpus moluccensis. Phytochemistry 2010, 71, 1917–1924. [Google Scholar] [CrossRef] [PubMed]
- Khalijah, A.; Chong, S.L.; Khalit, M.; Morita, H.; Hirasawa, Y.; Takeya, K.; Thoisone, O.; Hadi, A.H.A. Erythrocarpines A-E, new cytotoxic limonoids from Chisocheton erythrocarpus. Bioorg. Med. Chem. 2007, 15, 5997–6002. [Google Scholar]
- Zhao, J.; Feng, J.; Tan, Z.; Liu, J.; Zhao, J.; Chen, R.; Xie, K.; Zhang, D.; Li, Y.; Yu, L.; et al. Stachybotrysins A−G, Phenylspirodrimane Derivatives from the Fungus Stachybotrys chartarum. J. Nat. Prod. 2017, 80, 1819–1826. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Ushitora, H. Trapping of carbamic acid species with (trimethylsilyl)diazomethane. Tetrahedron 2006, 62, 226–235. [Google Scholar] [CrossRef]
- D’yakonov, V.A.; Dzhemileva, L.U.; Tuktarova, R.A.; Ishmukhametova, S.R.; Yunusbaeva, M.M.; Ramazanov, I.R.; Dzhemilev, U.M. Novel Hybrid Molecules on the Basis of Steroids and (5Z,9Z)-Tetradeca-5,9-dienoic Acid: Synthesis, Anti-Cancer Studies and Human Topoisomerase I Inhibitory Activity. Anti-Cancer Agents Med. Chem. 2017, 17, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Uyanik, M.; Ishihara, K. Hypervalent iodine-mediated oxidation of alcohols. Chem. Commun. 2009, 16, 2086–2099. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Maddox, M.M.; Adhikari, S.; Bruhn, D.F.; Kumar, M.; Lee, R.E.; Hurdle, J.G.; Lee, R.E.; Sun, D.Q. Syntheses and evaluation of macrocyclic engelhardione analogs as antitubercular and antibacterial agents. J. Antibiot. 2013, 66, 319–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Liu, Z.; Mi, Z.; Li, X.; Jia, P.; Zhou, J.; Yin, X.; You, X.; Yu, L.; Guo, F.; et al. High-throughput assay to identify inhibitors of Vpu-mediated down-regulation of cell surface BST-2. Antivir. Res. 2011, 91, 321–329. [Google Scholar] [CrossRef] [PubMed]






| Position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 2 | 3.00 t (8.4) | 2.95 t (8.4) | 2.77 dd (10.8, 2.4) | 2.57 m | 2.78 dd (10.4, 1.6) |
| 3 | 4.71 d (10.0) | 4.84 d (10.0) | 5.14 d (10.8) | 4.82 d (10.4) | 5.27 d (10.8) |
| 5 | 2.93 s | 2.84 d (10.0) | 2.98 d (10.8) | 2.92 br s | 2.64 a |
| 6a | 4.56 s | 2.35 m | 2.42 dd (16.8, 2.0) | 4.59 s | 2.35 m |
| 6b | 2.35 m | 2.42 dd (16.8, 10.8) | 2.64 a | ||
| 9 | 2.24 m | 2.15 dd (12.2, 5.2) | 2.65 br s | 2.46 m | |
| 11α | 1.76 m | 1.62 a | 1.73 m | 1.73 m | 1.97 m |
| 11β | 1.76 m | 1.77 m | 1.79 m | 1.73 m | 2.23 m |
| 12α | 1.52 m | 1.45 m | 1.11 m | 1.20 m | 1.51 m |
| 12β | 1.71 m | 1.62 a | 1.62 m | 1.62 m | 1.51 m |
| 14 | 2.31 br s | 2.30 br s | 2.52 mdd (11.6, 5.2) | ||
| 15α | 2.80 d (21.6) | 2.84 d (21.6) | 3.41 dd (21.2, 3.2) | 3.25 dt (21.6, 2.8) | 2.69 dd (16.0, 3.2) |
| 15β | 2.84 d (21.6) | 2.91 d (21.6) | 3.59 dd (21.2, 1.6) | 3.44 d (21.6) | 2.18 dd (16.0, 3.2) |
| 17 | 5.42 s | 5.55 s | 5.41 s | 5.33 s | 4.90 s |
| 18 | 0.98 s | 1.09 s | 1.08 s | 0.98 s | 0.78 s |
| 19 | 1.40 s | 1.07 s | 1.13 s | 1.38 s | 1.11 s |
| 21 | 7.50 br s | 7.74 br s | 7.53 br s | 7.44 br s | 7.44 br s |
| 22 | 6.38 br s | 6.45 br s | 6.46 br s | 6.38 br s | 6.41 br s |
| 23 | 7.44 br s | 7.41 br s | 7.42 br s | 7.42 br s | 7.42 br s |
| 28 | 0.77 s | 0.63 s | 0.56 s | 0.71 s | 0.70 s |
| Hpro-S-29 | 4.60 d (8.8) | 3.93 d (9.6) | 3.92 d (9.6) | 4.59 d (8.8) | 3.80 d (10.0) |
| Hpro-R-29 | 3.45 d (8.8) | 3.48 dd (9.6, 1.6) | 3.51 dd (9.6, 1.6) | 3.42 (8.8, 1.6) | 3.69 (10.0) |
| 30β | 5.28 d (6.8) | 5.31 d (6.8) | 4.51 d (2.4) | 2.24 m | 3.94 br s |
| 30α | 2.24 m | ||||
| 31 | 3.76 s | 3.69 s | 3.70 s | 3.84 s | 3.72 s |
| 3-Acyl | 3-Acyl | 3-Acyl | 3-Acyl | 3-Acyl | |
| 33 | 2.37 m | ||||
| 2.37 m | |||||
| 34 | 6.83 q (7.2) | 1.09 t (7.2) | 6.99 q (7.2) | 6.92 q (7.2) | 6.89 q (7.2) |
| 35 | 1.67 d (7.2) | 1.80 dd (7.2, 1.2) | 1.80 dd (7.2, 0.8) | 1.86 d (7.2) | |
| 36 | 1.79 s | 1.87 s | 1.86 s | 1.88 s |
| Position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | 96.9 qC | 96.9 qC | 97.4 qC | 97.1 qC | 97.4 qC |
| 2 | 45.3 CH | 45.1 CH | 48.2 CH | 43.6 CH | 46.2 CH |
| 3 | 76.2 CH | 75.2 CH | 75.2 CH | 78.5 CH | 74.8 CH |
| 4 | 36.6 qC | 36.2 qC | 37.1 qC | 36.6 qC | 36.8 qC |
| 5 | 39.6 CH | 35.0 CH | 34.5 CH | 39.3 CH | 40.6 CH |
| 6 | 72.5 CH | 31.9 CH2 | 32.1 CH2 | 73.1 CH | 31.9 CH2 |
| 7 | 176.1 qC | 173.9 qC | 174.2 qC | 175.8 qC | 174.3 qC |
| 8 | 138.3 qC | 138.4 qC | 131.3 qC | 130.6 qC | 127.1 qC |
| 9 | 48.9 CH | 47.8 CH | 38.7 CH | 43.9 CH | 139.2 qC |
| 10 | 42.1 qC | 41.6 qC | 45.1 qC | 44.7 qC | 43.6 qC |
| 11 | 20.4 CH2 | 19.4 CH2 | 17.7 CH2 | 18.2 CH2 | 21.1 CH2 |
| 12 | 34.6 CH2 | 34.5 CH2 | 29.4 CH2 | 30.3 CH2 | 29.4 CH2 |
| 13 | 36.7 qC | 36.9 qC | 38.2 qC | 37.8 qC | 35.3 qC |
| 14 | 44.8 CH | 45.0 CH | 134.3 qC | 129.0 qC | 36.8 CH |
| 15 | 29.6 CH2 | 30.1 CH2 | 32.3 CH2 | 33.1 CH2 | 32.2 CH2 |
| 16 | 169.3 qC | 170.1 qC | 169.5 qC | 169.8 qC | 172.7 qC |
| 17 | 76.6 CH | 77.0 CH | 80.7 CH | 81.6 CH | 81.3 CH |
| 18 | 21.2 CH3 | 21.9 CH3 | 17.4 CH3 | 18.0 CH3 | 20.2 CH3 |
| 19 | 14.9 CH3 | 14.4 CH3 | 14.2 CH3 | 15.1 CH3 | 14.3 CH3 |
| 20 | 121.5 qC | 120.8 qC | 120.7 qC | 121.1 qC | 121.2 qC |
| 21 | 140.3 CH | 141.8 CH | 141.6 CH | 140.9 CH | 140.4 CH |
| 22 | 109.3 CH | 109.7 CH | 109.9 CH | 109.8 CH | 109.7 CH |
| 23 | 143.1 CH | 143.0 CH | 142.9 CH | 143.1 CH | 143.2 CH |
| 28 | 15.8 CH3 | 14.8 CH3 | 15.6 CH3 | 16.4 CH3 | 18.2 CH3 |
| 29 | 70.1 CH2 | 68.0CH2 | 67.6 CH2 | 70.0 CH2 | 68.6 CH2 |
| 30 | 120.2 CH | 119.9 CH | 66.3 CH | 26.4 CH2 | 66.1 CH |
| 31 | 53.2 CH3 | 52.1 CH3 | 52.1 CH3 | 53.1 CH3 | 52.0 CH3 |
| 3-Acyl | 3-Acyl | 3-Acyl | 3-Acyl | 3-Acyl | |
| 32 | 167.2 qC | 174.5 qC | 167.6 qC | 167.5 qC | 167.6 qC |
| 33 | 127.8 qC | 27.2 CH2 | 128.8 qC | 128.9 qC | 128.4 qC |
| 34 | 138.7 CH | 8.8 CH3 | 139.6 CH | 138.7 CH | 139.2 CH |
| 35 | 14.6 CH3 | 14.6 CH3 | 14.6 CH3 | 14.8 CH3 | |
| 36 | 11.7 CH3 | 12.3 CH3 | 12.2 CH3 | 12.4 CH3 |
| cpd No. | Inhibition Rate (20.0 μM) (%) | IC50 Value (μM) | CC50 Value (μM) | cpd No. | Inhibition Rate (20.0 μM) (%) | IC50 Value (μM) | CC50 Value (μM) |
|---|---|---|---|---|---|---|---|
| 6 | 67.10 ± 3.04 | 19.23 ± 0.12 | > 100 | 8f | NA | NT | NT |
| 7 | NA | NT | NT | 8g | 50.84 ± 6.96 | 21.98 ± 4.65 | 86.76 ± 4.76 |
| 8 | NA | NT | NT | 8h | NA | NT | NT |
| 9 | NA | NT | NT | 8i | 99.95 ± 0.01 | 15.98 ± 6.87 | > 100 |
| 10 | NA | NT | NT | 9a | NA | NT | NT |
| 8a | NA | NT | NT | 9b | NA | NT | NT |
| 8b | 39.87 ± 2.83 | NT | NT | 10a | NA | NT | NT |
| 8c | 45.47 ± 6.84 | NT | NT | 10b | NA | NT | NT |
| 8d | 35.63 ± 8.85 | NT | NT | 10c | NA | NT | NT |
| 8e | 10.58 ± 3.55 | NT | NT | EFV | 88.78 ± 4.54 (4 nM) | NT | NT |
| cpd No. | Inhibition Rate (20.0 μM) (%) | IC50 Value (μM) | CC50 Value (μM) | cpd No. | Inhibition Rate (20.0 μM) (%) | IC50 Value (μM) | CC50 Value (μM) |
|---|---|---|---|---|---|---|---|
| 7 | 38.25 ± 9.67 | NT | NT | 8e | 43.42 ± 1.79 | NT | NT |
| 8 | NA | NT | NT | 8f | 44.94 ± 9.73 | NT | NT |
| 9 | NA | NT | NT | 8h | 48.15 ± 1.64 | NT | NT |
| 10 | NA | NT | NT | 9a | 44.72 ± 1.43 | NT | NT |
| 8a | 23.46 ± 2.42 | NT | NT | 10a | NA | NT | NT |
| 8b | 54.65 ± 1.43 | 17.93 ± 4.76 | 85.90 ± 4.65 | 10b | 63.40 ± 7.43 | 14.02 ± 3.54 | > 100 |
| 8c | 68.61 ± 0.99 | 15.87 ± 6.54 | 77.98 ± 4.65 | 10c | NA | NT | NT |
| 8d | 51.41 ± 1.54 | 22.78 ± 1.86 | 87.65 ± 4.76 | ribavirin | 87.54 ± 2.86 (60 μM) | NT | NT |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, J.-L.; Zou, X.-P.; Li, W.-S.; Shen, L.; Wu, J. Limonoids Containing a C1–O–C29 Moiety: Isolation, Structural Modification, and Antiviral Activity. Mar. Drugs 2018, 16, 434. https://doi.org/10.3390/md16110434
Ren J-L, Zou X-P, Li W-S, Shen L, Wu J. Limonoids Containing a C1–O–C29 Moiety: Isolation, Structural Modification, and Antiviral Activity. Marine Drugs. 2018; 16(11):434. https://doi.org/10.3390/md16110434
Chicago/Turabian StyleRen, Jing-Ling, Xiao-Peng Zou, Wan-Shan Li, Li Shen, and Jun Wu. 2018. "Limonoids Containing a C1–O–C29 Moiety: Isolation, Structural Modification, and Antiviral Activity" Marine Drugs 16, no. 11: 434. https://doi.org/10.3390/md16110434